Abstract
A Luminex xTAG-based assay for plant-infecting tospoviruses was developed. The test enables the detection of tospoviruses in general and the differentiation of the four important member species of this genus: Tomato spotted wilt virus, Impatiens necrotic spot virus, the proposed ‘Capsicum chlorosis virus’ and Watermelon silver mottle virus. The generic tospovirus primers used in this method are also applicable for detection of tospoviruses by basic RT-PCR. We also describe an economic alternative method for the distinction of the four tospoviruses mentioned and of additional member viruses, based on a restriction fragment length polymorphism (RFLP). The sophisticated Luminex xTAG technology allows the simultaneous detection of various targets. This study is part of a project that aims to develop a method for the simultaneous detection of various plant pathogens (viral, bacterial and fungal) in plant material.
Keywords: Cucumber Mosaic Virus, Tomato Yellow Leaf Curl Virus, Tomato Spotted Wilt Virus, Median Fluorescence Intensity, Iris Yellow Spot Virus
Introduction
The genus Tospovirus comprises all plant-infecting viruses of the family Bunyaviridae. The other four genera of this family (Orthobunyavirus, Phlebovirus, Nairovirus and Hantavirus) contain animal infecting viruses. The genus Tospovirus obtained its name from the type species Tomato spotted wilt virus and contains 11 approved species and 20 tentative species [1–3]. These viruses have quasi-spherical particles, with a diameter of 80-120 nm, enveloped by a host-derived membrane. The two glycoproteins Gn and Gc are embedded in this membrane. Particles contain a tripartite single-stranded RNA genome with negative- or ambi-sense polarity. The three RNAs differ in size and are thus called large (L), medium (M) and small (S). All three RNAs are incorporated in one particle but independently packaged by many copies of the nucleoprotein and a few copies of the viral RNA-dependent RNA polymerase [4, 5].
Tospoviruses have a large host range: tomato spotted wilt virus (TSWV) infects for example 1,090 plant species belonging to 85 families, including many economically important crop plants and numerous weed species [6]. On the other hand, other tospoviral species have a more restricted host range. Some of the members, such as TSWV, impatiens necrotic spot virus (INSV) and iris yellow spot virus (IYSV), are found worldwide, while others are restricted to certain regions: for example watermelon silver mottle virus (WSMoV) is restricted to Asia, capsicum chlorosis virus (CaCV) to Asia and Australasia and polygonum ringspot virus (PolRSV) to Europe [7]. The same applies to the thrips vector species (insect order: Thysanoptera): some have a worldwide distribution like Frankliniella occidentalis [8], while others are restricted to single countries: for example Thrips setosus can be found only in Japan [9]. Both species of thrips are vectors of TSWV [10]. Overall, fifteen species of thrips transmit tospoviruses in a persistent and propagative manner [10] and are critical for their epidemiology [7]. The host range and geographical distribution of the established tospovirus species have increased in recent years [7]. Additionally, new species belonging to the Tospovirus genus have been proposed in recent years, such as ‘Alstroemeria necrotic streak virus’ [11], ‘Pepper necrotic spot virus’ [12] and ‘Tomato necrotic spot virus’ [13].
Based on the large loss of produce and economic damage caused, tospoviruses are thought to be the most devastating plant viruses [14, 15]. This makes the diagnosis of tospoviral infections an important issue, especially for plant protection services trying to confine further spread of associated diseases. ELISA, RT-PCR and quantitative or real time RT-qPCR can be used to detect tospoviruses and to discriminate different species. Because of its robustness and sensitivity, ELISA is often the standard method of choice for plant virus diagnostics [16]. Tospoviruses can be serologically detected by polyclonal antibodies against the nucleocapsid protein and distinguished into four major serogroups with TSWV, WSMoV, IYSV and groundnut yellow spot virus (GYSV) being the type species [17, 18]. Some tospoviruses have no clear serological relationship to the serogroups and are seen as distinct mono-serotypes [19]. In most cases, monoclonal antibodies can also be used to classify tospoviral species in serogroups [19], but some tospoviruses with high amino acid sequence similarity such as CaCV, WSMoV, groundnut bud necrosis virus (GBNV) and watermelon bud necrosis virus (WBNV), are difficult to distinguish even with monoclonal antibodies [20, 21]. This cross-reactivity has also been described when using commercial antibodies or ELISA tests supplied by the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (DSMZ; Braunschweig, Germany), Agdia (Elkhart, USA) and LOEWE Biochemica (Sauerlach, Germany). For TSWV antibodies, possible cross-reactivity with groundnut ringspot virus (GRSV), tomato chlorotic spot virus (TCSV) and chrysanthemum stem necrosis virus (CSNV) are known. This cross-reactivity among species complicates distinction of different tospoviruses by ELISA. Additionally, an ELISA test has to be performed independently for every species, since multiplexing is not possible. Multiplex RT-PCRs can be a solution for this and have been described for the detection of TSWV, INSV, CSNV, IYSV and CaCV [22] as well as for detection of TSWV, melon yellow spot virus (MYSV), WSMoV, INSV and IYSV [23].
In this study a new method for plant virus diagnosis is described using the Luminex xTAG technology to test for tospoviruses in general and for the four species TSWV, WSMoV, INSV and CaCV. Virus samples of 12 tospovirus isolates from eight different species were obtained from the DSMZ. The nucleic acid-based assay platform of the Luminex xTAG technology allows the simultaneous detection of theoretically up to 500 analytes in one sample. Tospoviral RNA is transcribed into cDNA and amplified in a first RT-PCR. The products are then subjected to a target specific primer extension (TSPE) reaction, for which tagged primers and biotinylated dCTP are used. The primer tags allow the hybridization of TSPE products to complementary anti-tags coupled to Luminex MagPlex-TAG Microspheres. These paramagnetic polystyrene “beads” are filled with a mix of two to three fluorescent dyes at different ratios, enabling their later identification through excitation and measured fluorescence and thus the identification of bound TSPE products. The presence of hybridized TSPE products is revealed through the binding of streptavidin-R-phycoerythrin to the incorporated biotin, its excitation and detection of the resulting fluorescence. The test procedure is described in detail by van der Vlugt et al. [24]. This technique has been used in human medicine for diagnosis of respiratory viruses such as influenza and coronaviruses, among others [25]. In recent years, this technique has been adapted for the diagnostics of plant pathogens. Van Brunschot et al. detected and distinguished different begomoviruses and their whitefly vectors [26] and different pospiviroids [27] applying this method. Lim et al. [28] used it for the simultaneous detection of three lily-infecting viruses. Together, these studies demonstrate the advantage of this approach which allows combination of tests for various pathogens to detect the most important diseases of a particular crop. Our work should contribute to this end, as part of a project for the development of a simultaneous detection method for various viral, bacterial and fungal plant pathogens. Additionally, this study describes a more economical approach for generic tospovirus detection by RT-PCR and for the differentiation of tospoviral species by restriction fragment length polymorphism (RFLP).
Materials and methods
Virus specimens
Infected, dried plant material of 12 tospoviral isolates from eight different species was obtained from the DSMZ including single isolates of alstroemeria necrotic streak virus (ANSV), CaCV, GRSV, IYSV, TCSV and WSMoV as well as three isolates each of INSV and TSWV. Information about the viral origin (host plant, country of origin and provider) was supplied by the DSMZ (Table 1) for most isolates. All 12 isolates were mechanically inoculated on Nicotiana benthamiana plants using phosphate buffer (0.05 M KH2PO4, 0.05 M Na2HPO4, 1 mM EDTA, 5 mM Na-DIECA) with a spatula tip of celite and charcoal. Inoculated plants were grown in a greenhouse and monitored for symptom expression. Leaves of plants showing systemic symptoms (after one to two weeks) were used for RNA extraction.
Table 1.
Tospovirus species and isolates from the DSMZ with information about the original host plant, the country of origin and the provider as supplied by the DSMZ
| Tospovirus | Isolate | Host plant | Country of origin | Provider |
|---|---|---|---|---|
| Alstroemeria necrotic streak virus (ANSV) | PV-1027 | Alstroemeria sp. | Colombia | R. Kormelink |
| Capsicum chlorosis virus (CaCV) | PV-0864 | Solanum lycopersicum | Thailand | E. Maiss |
| Groundnut ringspot virus (GRSV) | PV-0205 | Arachis hypogaea | South Africa | G. Pietersen |
| Impatiens necrotic spot virus (INSV) | PV-0280 | Hippeastrum sp. | USA | O.W. Barnett |
| INSV | PV-0281 | Anemone coronaria | Germany | J. Dalchow |
| INSV | PV-0485 | Gloxinia sp. | unknown | P. Roggero |
| Iris yellow spot virus (IYSV) | PV-0528 | Allium ampeloprasum | unknown | J.T.J. Verhoeven |
| Tomato chlorotic spot virus (TCSV) | PV-0391 | Capsicum annuum | Brazil | O. Lovisolo |
| Tomato spotted wilt virus (TSWV) | PV-0182 | Nicotiana tabacum | Bulgaria | M. Jankulova |
| TSWV | PV-0204 | Impatiens New Guinea Hybrid | unknown | D.-E. Lesemann |
| TSWV | PV-0393 | Nicotiana tabacum | Bulgaria | M. Jankulova |
| Watermelon silver mottle virus (WSMoV) | PV-0283 | Solanum lycopersicum | Taiwan | S.K. Green |
Nucleic acid extraction
Total RNA was extracted once from infected, dried plant material, twice from infected N. benthamiana plants and three times from healthy N. benthamiana plants. The RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used, following the manufacturer’s instructions.
Primer design
Primers for general tospovirus detection, for pre-amplification and for TSPE reactions were designed on the basis of alignments of nucleotide sequences. Sequences of segment M were retrieved from GenBank (National Center for Biotechnology Information), imported into CLC Main Workbench (CLC bio, Aarhus, Denmark) and aligned. For the general detection primers (Tospo_GENs/as), the pre-amplification primers (Tospo_OUTs/Tospo_GENas) and the generic TSPE primers (tTospo_GENs/as), the alignments were analyzed for conserved regions and corresponding sequences were chosen for the primers. Some degenerate bases were inserted into primers sequences. For the species-specific TSPE primers tTSWVs/as, tINSVs/as, tWSMoVs/as and tCaCVs/as, conserved regions for each of the species were identified in the alignments and relevant sequences used for the primers. As internal control primers we used the pre-amplification primers (Nad5s/as) for the NADH dehydrogenase subunit 5 gene (nad5) from Menzel et al. [29] and the TSPE primer (tNad5) for nad5 from van Brunschot et al. [26] and adapted from Botermans et al. [30]. The tag sequences were added to all TSPE primers by Luminex (Austin, USA) and were complementary to anti-tags on the microspheres’ surfaces. The primers were synthesized by Eurofins Scientific (Luxembourg). Table 2 displays their characteristics.
Table 2.
Characteristics of oligonucleotide primers used in this study
| Name | Sequence (5′-3′) | Length [nts] | Fragment size [bp] | Corresponding microsphere | |
|---|---|---|---|---|---|
| Tospo_GENs | TCHTNCCAACHTGGRAYAG | 19 | 420 | ||
| Tospo_GENas | TGCADGCYTCAATNAADGC | 19 | 480 | ||
| Tospo_OUTs | TVACHAAYTGGAARAATGA | 19 | |||
| Nad5s | GATGCTTCTTGGGGCTTCTTGTT | 23 | 180 | ||
| Nad5as | CTCCAGTCACCAACATTGGCATAA | 24 | |||
| tTospo_GENs | CTTAAACTCTACTTACTTCTAATT-TCHTNCCAACHTGGRAYAG | 43 | 420 | MTAG-A056 | |
| tTospo_GENas | CATAAATCTTCTCATTCTAACAAA-TGCADGCYTCAATNAADGC | 43 | MTAG-A075 | ||
| tTSWVs | ACAAATATCTAACTACTATCACAA-AACCCTACAGGGAAAC | 40 | 180 | MTAG-A039 | |
| tTSWVas | TTAACAACTTATACAAACACAAAC-CTGCACATCAAATGC | 39 | MTAG-A053 | ||
| tINSVs | ATACTTTACAAACAAATAACACAC-ACCAAGATAATTAAGATACA | 44 | 260 | MTAG-A019 | |
| tINSVas | ATCTCAATTACAATAACACACAAA-AAGCTGAACACAATTC | 40 | MTAG-A067 | ||
| tWSMoVs | CACTACACATTTATCATAACAAAT-GTCAGTTTCACTATAAGC | 42 | 260 | MTAG-A042 | |
| tWSMoVas | CTATCATTTATCTCTTTCTCAATT-TAAGTTGCATGCACTG | 40 | MTAG-A072 | ||
| tCaCVs | CTAAATCACATACTTAACAACAAA-GTCAGCTTCACTATAAAT | 42 | 260 | MTAG-A063 | |
| tCaCVas | TTCAATTCAAATCAAACACATCAT-TTGAGTTGCATGCAGTA | 41 | MTAG-A064 | ||
| tNad5 | AACTTTCTCTCTCTATTCTTATTT-AGGATCCGCATAGCCCTCGATTTATGTG | 52 | MTAG-A043 | ||
TSPE primer names are preceded by the letter t, their tag sequences are italicized and their corresponding Luminex MagPlex-TAG Microspheres with bead addresses are listed
Pre-amplification RT-PCR
Viral RNA was transcribed into cDNA and then amplified using the Access RT-PCR System kit (Promega, Fitchburg, USA), using the concentrations specified by the manufacturer in a volume of 25 µl, in covered 96-well Multiply PCR plates (Sarstedt, Nuembrecht, Germany). The following incubations were carried out: 45 min at 45 °C, 2 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 47 °C and 30 s at 68 °C, followed by a final extension of 7 min at 68 °C. The internal control primers (Nad5s/as) and the degenerate primers (Tospo_OUTs/Tospo_GENas) were used, enabling the production of a 180 bp fragment for all plant samples and a 480 bp fragment for all tospoviruses. After RT-PCR, 5 µl of each product were stained with 1 µl of loading buffer (37.5% glycerol, 0.2% bromophenol blue, 125X GelRed [Biotium, Hayward, USA]) and loaded on an agarose gel (1%) to visualize the expected fragments.
TSPE reaction
The pre-amplification products were directly used for a multiplex TSPE reaction. Primers were extended and biotin-14-dCTP (Thermo Fisher Scientific, Waltham, USA), instead of normal dCTP, was incorporated, alongside with the remaining unmodified nucleotides (dATP, dGTP, dTTP; Thermo Fisher Scientific). A set of eleven primers was used: two generic primers, to detect all tospoviruses (tTospo_GENs/as), eight specific primers, to identify four tospovirus species (tTSWVs/as, tINSVs/as, tWSMoVs/as and tCaCVs/as) and one plant internal control primer (tNad5) for nad5. TSPE mixes of 20 µl containing 5 µl of the pre-amplification products were prepared (0.75 U Platinum GenoTYPE Tsp DNA polymerase [Thermo Fisher Scientific], 5 µM biotin-14-dCTP, 5 µM each of normal dATP, dGTP and dTTP, 1.5 mM MgCl2, 25 nM of each primer, 1X PCR reaction buffer [Thermo Fisher Scientific] and sterile water up to 20 µl). The reaction mix was incubated under the following conditions in covered 96-well Multiply PCR plates: one cycle of 2 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 45 °C and 1 min at 72 °C, followed by a final extension of 5 min at 72 °C.
Microsphere hybridization
The TSPE products were hybridized to the corresponding beads. A mix of the eleven Luminex MagPlex-TAG Microspheres (MTAG-A075, MTAG-A072, MTAG-A067, MTAG-A064, MTAG-A063, MTAG-A056, MTAG-A053, MTAG-A043, MTAG-A042, MTAG-A039 and MTAG-A019) was prepared (0.5 µl of each bead per reaction containing 1,250 beads) in 2X Tm hybridization buffer (0.4 M NaCl, 0.2 M Tris, 0.16% Triton X-100, pH 8.0; 25 µl per reaction) and double distilled and deionized water (13.5 µl per reaction). The mixture was split into 96-well polycarbonate microplates (Corning, Corning, USA) and 5 µl of the TSPE products were added to each well, for a final volume of 50 µl with 25 microspheres per microliter of hybridization mixture. Interactions between the complementary tag sequences of the TSPE primers and anti-tag sequences coupled to the microspheres allowed the hybridization of TSPE products to the beads. To facilitate this, the mixtures were first denatured at 96 °C for 90 s and then incubated at 37 °C for 30 min.
Luminex assay
After hybridization the mixtures were moved to Cellstar 96-well cell culture plates (Greiner Bio-One, Kremsmuenster, Austria) and the microspheres were pelleted on a magnetic separator for 1 min. The supernatants were discarded and the beads resuspended and washed twice in 80 µl 1X Tm hybridization buffer per well. The supernatants were removed again and 80 µl of 1X Tm hybridization buffer containing streptavidin-R-phycoerythrin (2 µg/ml; Thermo Fisher Scientific) were added to each well. The plates were protected from light and incubated on a shaker (600 rpm) at room temperature for 15 min. After pelleting the microspheres again, the supernatants were discarded and the microspheres resuspended in 90 µl of 1X Tm hybridization buffer per well. Finally, 70 µl per sample were analyzed in a Luminex 200 System with the xPONENT Software (Version 3.0; Luminex). A red laser (635 nm) excited the bead dyes and a green laser (532 nm) excited the R-phycoerythrin, bound via streptavidin and biotin to the TSPE products. The fluorescence of the R-phycoerythrin was recorded as the median fluorescence intensity (MFI) signal, allowing identification of samples containing amplified plant or tospoviral nucleic acids. Identification of these nucleic acids could be achieved by the fluorescence of the bead dyes, which are unique for each microsphere with its specific anti-tag. Three independent measurements were performed with the three RNA extractions described above.
RT-PCR and RFLP
For the general detection of tospoviruses, RNA from infected plant material was transcribed into cDNA by incubating 4 µl of water, 2 µl of RNA, 1 µl of primer Tospo_GENas (10 µM) and 0.5 µl of dNTP mix (10 mM each) at 99 °C for 3 min. The reaction mix was then cooled on ice and 0.5 µl of RevertAid reverse transcriptase (20 U/µl; Thermo Fischer Scientific) and 2 µl of 5X reaction buffer (RevertAid; Thermo Fisher Scientific), were added. The mixture was then incubated at 42 °C for 60 min. The cDNA was amplified in a reaction mix consisting of 10 µl of Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific), 6 µl of water, 2 µl of cDNA, 1 µl of primer Tospo_GENs and 1 µl of primer Tospo_GENas and was incubated under the following conditions: one cycle for 15 s at 98 °C, 34 cycles of 5 s at 98 °C, 5 s at 52 °C and 15 s at 72 °C and a final extension of 5 min at 72 °C. PCR products were analyzed by gel electrophoresis on an agarose gel. For distinction of tospoviruses by RFLP, an RT-PCR was performed like above and the products were digested with the restriction enzymes HinfI (Thermo Fisher Scientific) and BclI (New England Biolabs, Ipswich, USA). For this, 20 µl of each PCR product were mixed with 2 µl of HinfI (10 U/µl), 2 µl of BclI (10 U/µl), 3 µl of 10X FastDigest buffer (Thermo Fisher Scientific) and 3 µl of water and incubated at 37 °C for one hour. RFLP reactions were visualized using gel electrophoresis.
Results
Luminex assay for tospovirus detection and distinction
We verified by conventional RT-PCR experiments that the pre-amplification and generic primers worked with tospovirus RNA and that the species-specific primers were specific for the corresponding virus species (data not shown). After pre-amplification with the four primers Tospo_OUTs/Tospo_GENas and Nad5s/as, the RT-PCR products were used in TSPE reactions with all eleven TSPE primers (tTospo_GENs/as, tTSWVs/as, tINSVs/as, tWSMoVs/as, tCaCVs/as and tNad5). TSPE products were hybridized to the corresponding eleven MagPlex-TAG Microspheres listed in Table 2 and the hybridization mix was analyzed. Three independent Luminex tests were conducted to verify the reliability of the method. Every test included new RNA extractions, pre-amplifications, TSPE reactions, hybridizations and Luminex measurements. The mean values of the MFIs of the three Luminex measurements were determined and plotted in Fig. 1.
Fig. 1.
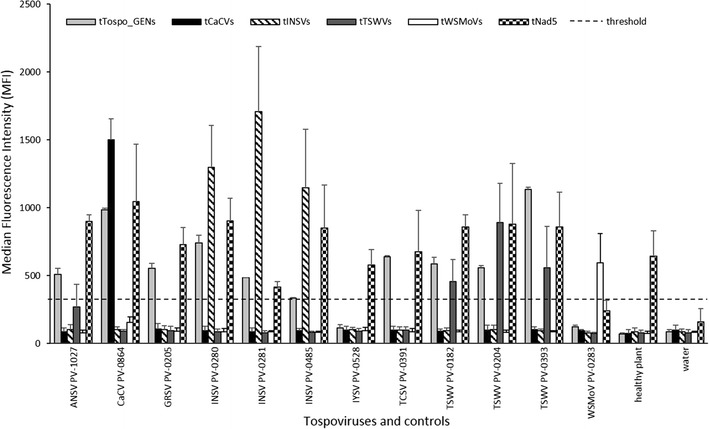
Results of three independent measurements of the Luminex assay. The tests were conducted against tospoviruses in general, against the tospoviral species CaCV, INSV, TSWV and WSMoV as well as against a plant internal control. Plant material isolated from a series of plants, each infected with one of the 12 tospoviral isolates, healthy plant material and a water control were used in each test. The MFI values were measured and the mean values of the three measurements were plotted as well as the standard deviations. The threshold for all tests was set at twice the mean MFI value of the water control of the tNad5 test (dashed line)
The generic primer tTospo_GENs detected most tospoviruses, except IYSV and WSMoV. The positive signal ranged from 333 for one INSV isolate to 1,135 for one TSWV isolate. IYSV- and WSMoV-infected plant material produced a signal of 117 and 123, respectively, not exceeding the threshold set at 323 (light grey columns in Fig. 1). The internal control TSPE primer (tNad5) detected the nad5 gene in previously amplified plant material in almost all samples, with MFIs ranging from 415 for one INSV isolate to 1,048 for CaCV. Only in the WSMoV sample the threshold was not reached, with a value of 239 (checkered columns in Fig. 1). Each species-specific sense TSPE primer correctly identified the target virus species it was designed to detect. The tCaCVs primer produced a high signal of 1,499 in the CaCV infected samples but no signal was detected in any other sample or control (black columns in Fig. 1). The tINSVs primer lead to high MFI values of 1,148 to 1,708 only in the samples infected with the three INSV isolates (diagonally striped columns in Fig. 1). The signal induced by the tWSMoVs primer was slightly lower (594) and also restricted to samples infected with WSMoV (white columns in Fig. 1). The MFI values generated by the tTSWVs primer were also slightly lower (455-891) and restricted to samples containing the three TSWV isolates tested. In this case, a slight reaction (MFI of 268) also occurred in plants infected with ANSV (dark grey columns in Fig. 1). The five antisense TSPE primers (tTospo_GENas, tTSWVas, tINSVas, tWSMoVas and tCaCVas) were tested as alternatives for the sense primers, but they did not generate satisfactory results in Luminex tests for tospovirus detection and distinction (data not shown). As a threshold for all tests, an MFI value twice the MFI value of the water control from the tNad5 test was chosen, because this sample showed the highest MFI value of all healthy plant and water controls. The values of the other tests were comparable, but were left out of the figure for the sake of clarity (dashed line in Fig. 1).
Generic and specific tospovirus detection by RT-PCR
The generic tospovirus primers Tospo_GENs/as (without tags) allowed the detection of all eight tospoviruses and of all three isolates of INSV and TSWV tested in RT-PCR experiments. The expected fragment of about 420 bp was produced when using RNA extracted from infected plants. The primers generated no band when using RNA from healthy plants or from plants infected with other viruses like cucumber mosaic virus (CMV), plum pox virus (PPV) and pepper mild mottle virus (PMMoV) (Fig. 2), demonstrating the specificity of the primers.
Fig. 2.
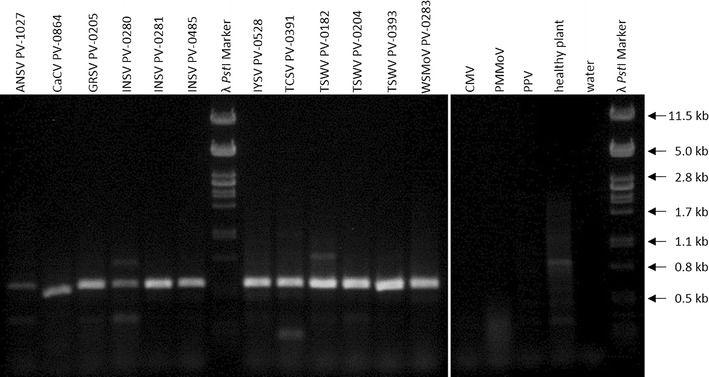
Results of a RT-PCR with RNA from plants infected with 12 tospovirus isolates, CMV, PMMoV and PPV as well as RNA from healthy plants and a water control using primers Tospo_GENs/as after gel electrophoresis. Enterobacteria phage λ DNA digested with PstI was used as molecular-weight size marker. DNA fragments appear larger than the expected 420 bp, because the DNA stain GelRed changes the migration speed of DNA, depending on the concentrations of DNA and GelRed [40]
The four species-specific primer pairs (without tags) also worked in normal RT-PCRs for the identification of these species (data not shown). These primers led to the production of the expected fragments only when RNA from plant material infected with the corresponding virus was used. For TSWV the specific fragment was 180 bp in size and for CaCV, INSV and WSMoV 260 bp. Primer sequences can be deduced from Table 2 omitting the tag sequences in italics.
RFLP distinction of tospoviruses
Most of the examined tospovirus species could be distinguished by a RFLP analysis using the restriction enzymes HinfI and BclI after RT-PCR with primers Tospo_GENs/as. The PCR products of the tospoviruses have different restriction enzyme recognition sites which leads to a distinct pattern for most tospoviruses after gel electrophoresis. RFLP analysis was first performed in silico using the software CLC Main Workbench and tospoviral segment M sequences from GenBank, to predict fragment sizes. After RT-PCR, digestion and RFLP analysis, the predicted fragments were observed for most viruses: for TSWV these were 325 and 90 bp in size, for INSV they were 260, 90 and 70 bp, for WSMoV 290, 50, 30 and 20 bp, for CaCV 280, 80 and 30 bp, for GRSV 320 and 90 bp and for TCSV they were 140, 90 and 50 bp. For IYSV, fragments of 310, 80 and 20 bp were detected, but a predicted 10 bp fragment was not detected. For ANSV no predictions could be made in silico because for this virus only a partial segment S sequence (containing the nucleocapsid protein gene) is available. Nevertheless, the RFLP pattern for the ANSV was determined and fragments of about 320 and 90 bp were detected. Therefore, the RFLP pattern of ANSV was similar to that of GRSV and TSWV, and these three viruses could not be clearly differentiated. CaCV, INSV, IYSV, TCSV and WSMoV could be distinguished from these viruses and from one another (Fig. 3).
Fig. 3.
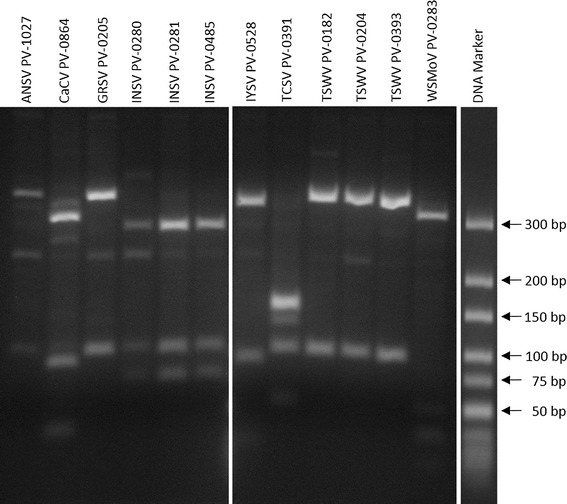
Results of a RFLP after gel electrophoresis. RNA from plants infected with 12 tospovirus isolates was transcribed and amplified in a RT-PCR with primers Tospo_GENs/as. PCR products were then digested using HinfI and BclI. The O’GeneRuler Ultra Low Range DNA Ladder (Thermo Fisher Scientific) was used as molecular-weight size marker. The figure was assembled from two gels
Discussion
A molecular assay for the detection of tospoviruses in general and for viruses from the four species belonging to this genus (TSWV, INSV, CaCV and WSMoV), using the Luminex xTAG technology, was successfully developed. The generic tospovirus test with primer tTospo_GENs detected six of the eight tested species. It failed to prove the presence of IYSV and WSMoV. However, the same primer without its tag, in combination with an antisense primer, detected these two viruses in RT-PCR experiments. These RT-PCR results suggest that with further optimization, the generic tospovirus Luminex test could be improved to detect all eight species. The failure to detect IYSV and WSMoV might be a sensitivity problem of the generic primer in the tospovirus Luminex test. The virus concentration in these samples could be too low for detection, as the TSPE reaction only linearly amplifies the target, while a conventional PCR leads to an exponential amplification. Alternatively, primers might not bind perfectly to target sequences or the tag sequences of the TSPE primers might interact non-specifically with plant or viral sequences. In this case, adaptations of the primer or tag sequences might solve the problem. The plant internal control primer tNad5 detected its target except in the case of WSMoV samples. This result probably points to a low nucleic acid concentration in the WSMoV samples preventing successful detection of the internal control due to a sensitivity problem in this Luminex assay. By increasing the sample number, the low nucleic acid concentration and low MFI values of single samples in the hybridization mix would probably be corrected, leading to exceedance of the threshold. The species-specific tests using the primers tCaCVs, tINSVs, tTSWVs and tWSMoVs were specific for the viruses they were designed to detect. In the case of tINSVs and tTSWVs, all three isolates of each of these two viruses were successfully detected.
This study is one of only a few employing the Luminex xTAG technology for the detection of plant pathogens and the first for tospoviruses. So far it has been applied to screen for begomoviruses and pospiviroids [26, 27], for lily mottle virus (LMoV) and lily symptomless virus (LSV) in lily plants [28] and for CMV and its two subgroups [31]. The related Luminex xMAP technology utilizing coupled antibodies instead of oligonucleotides has been used to identify potato virus X (PVX), potato virus Y (PVY) and potato leaf roll virus (PLRV) [32] as well as plum pox virus (PPV) [33] in plant material. However, antibodies cross reactivity between different tospoviral species has been described by researchers [2, 20] and antisera suppliers (DSMZ, Agdia and LOEWE Biochemica). This may be a problem in the antibody-based Luminex xMAP test for the differentiation of tospovirus species, and hence a nucleic acid based array is thought to be advantageous. An advantage of both these assay formats is their multiplexing capability and their ability to simultaneously detect various diseases in plant samples. The study of Lim et al. [28] is an example of this since they used a Luminex xTAG assay for the detection of the three viruses (CMV, LMoV and LSV) infecting lily plants. The standard method for virus detection (ELISA) lacks this potential and is quite labor- and time-intensive as it only allows to test for one virus at a time. Charlermroj et al. [34] have created a multiplex antibody array similar to an ELISA for the simultaneous detection of the three viruses MYSV, WSMoV and chilli veinal mottle virus (ChiVMV) as well as of the fruit blotch bacterium Acidovorax avenae subsp. citrulli. All four pathogens were immobilized by capture antibodies specific to the four pathogens in each well at preassigned positions, detected by fluorescently conjugated secondary antibodies and identified by their position in the microwells. Such multiplexed antibody array technologies are still under development and are unlikely to be used for routine plant pathogen detection. A mixed detection method combining RT-PCR and ELISA was developed and applied for the detection of the four tospoviruses CaCV, MYSV, tomato necrotic ringspot virus (TNRV) and WSMoV. Using this technique, the RNA is first transcribed and amplified by RT-PCR using degenerate primers and digoxigenin (DIG) labelled dUTP, then the PCR products are hybridized to four species-specific biotinylated probes in streptavidin-coated microtiter wells and finally the labelled and hybridized PCR products are detected by ELISA using a peroxidase-conjugated anti-DIG antibody [35].
Our Luminex xTAG test for tospoviruses could be combined with already existing and prospective tests for plant pathogens to create assays that can identify crops’ most important diseases, similar to the respiratory virus panel test developed by Mahony et al. [25] that screens for 20 different human respiratory viruses and their subtypes. For example, for tomato crops (Solanum lycopersicum) we could combine the tests for TSWV, CaCV and INSV from this study, for tomato yellow leaf curl virus (TYLCV) from van Brunschot et al. [26], for tomato apical stunt viroid (TASVd), tomato chlorotic dwarf viroid (TCDVd) and tomato planta macho viroid (TPMVd) from van Brunschot et al. [27] as well as the test for CMV from Lim et al. [28] or from Bald-Blume et al. [31]. Further tests could be added to cover the most important of the 136 viruses infecting tomatoes [36, 37] and also bacterial and fungal pathogens.
A more economical alternative for generic tospovirus detection and species distinction was successfully developed in this study. The primer Tospo_GENs/as detected RNA of all eight examined tospoviruses in RT-PCRs but not of viruses from other genera. This is one of the best coverages of tospoviral species obtained by one primer pair so far. Hassani-Mehraban et al. [2] reported an overview of universal, degenerate and multiplex primers for tospovirus detection from eleven studies (details of covered species are reported in Table 4 of [2]). Chen et al. [38] designed two degenerate primer pairs that transcribed and amplified RNA of 12 species. One of the pairs binds to segment L and the other to the NSm gene of segment M. Their antisense primer gM870c for segment M partially coincides with our Tospo_GENas primer. Since our primer is more degenerated, our primers might also identify RNA from the additional tospoviral species GBNV, calla lily chlorotic spot virus (CCSV), MYSV, WBNV and tomato yellow ring virus (TYRV) tested by Chen et al. [38], although we have not tested this. Hassani-Mehraban et al. [2] describe six primer pairs and RT-PCRs that cover 20 assigned and tentative tospoviral species and classify them into six subgroups.
Also, the four species-specific primer pairs (without tags) can be used in RT-PCRs for the identification of these species. They lead to the amplification of nucleic acids only of the corresponding species and of all three isolates tested from either TSWV or INSV. They cannot be used for multiplex RT-PCRs, because the CaCV, INSV and WSMoV specific primers lead to fragments of the same size. So a distinction of these species would not be possible by a plain multiplex RT-PCR. The fragments of the generic tospovirus primers and the TSWV specific primers can be distinguished from these primers and from one another, and could be combined with one of the other four primer pairs for a clear multiplex RT-PCR. Kuwabara et al. [22] described such a multiplex RT-PCR for the detection of the five tospoviruses TSWV, INSV, CSNV, IYSV and CaCV.
Alternatively, RT-PCR products of the different tospovirus species generated with the generic primers Tospo_GENs/as could be differentiated by RFLP using the restriction enzymes HinfI and BclI. The five species CaCV, IYSV, INSV, TCSV and WSMoV were clearly distinguished. The additional three species ANSV, GRSV and TSWV could be discriminated from the rest but not clearly from each other. Chu et al. [39] used this method to detect and differentiate the five tospoviruses TSWV, GRSV, INSV, WSMoV and peanut chlorotic fan-spot virus (PCFV) using primers binding to segment L and the restriction enzyme XbaI.
The three methods described in this study for tospovirus detection and species differentiation (Luminex xTAG, RT-PCR and RFLP) are comparable in detection efficiency. The Luminex xTAG assay seems to be less sensitive than the other two methods. However, an advantage of the Luminex xTAG technology is the high multiplexing potential leading to a reduction in labor when testing for several possible tospoviruses and other plant pathogens.
Acknowledgements
We thank Prof. Dr. med. Rainer Blasczyk and Prof. Dr. med. Stephan Immenschuh of the Institute for Transfusion Medicine of Hannover Medical School (MHH) for providing their Luminex 200 system and Dr. Eva Zilian, Susanne Aufderbeck and Stefanie Vahlsing of the same institute for their support.
Compliance with ethical standards
Funding
This study was partly funded by the EU Interreg IV A program Gezonde Kas (healthy greenhouse).
Conflict of interest
The authors declare that there are no conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB. Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. London: Elsevier Academic Press; 2012. pp. 725–741. [Google Scholar]
- 2.Hassani-Mehraban A, Westenberg M, Verhoeven JTJ, van de Vossenberg BTLH, Kormelink R, Roenhorst JW. Generic RT-PCR tests for detection and identification of tospoviruses. J Virol Methods. 2016;233:89–96. doi: 10.1016/j.jviromet.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 3.International Committee on Taxonomy of Viruses (2015) Master Species List #30. http://www.ictvonline.org/virusTaxonomy.asp. Accessed 14 Jul 2016
- 4.Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni A-L. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 2011;162(1–2):184–202. doi: 10.1016/j.virusres.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Turina M, Tavella L, Ciuffo M. Tospoviruses in the Mediterranean area. In: Loebenstein G, Lecoq H, editors. Advances in virus research: viruses and virus diseases of vegetables in the Mediterranean basin. San Diego: Elsevier Academic Press; 2012. pp. 403–437. [DOI] [PubMed] [Google Scholar]
- 6.Parrella G, Gognalons P, Gebre-Selassiè K, Vovlas C, Marchoux G. An update of the host range of tomato spotted wilt virus. J Plant Pathol. 2003;85(4):227–264. [Google Scholar]
- 7.Pappu HR, Jones RAC, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141(2):219–236. doi: 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Kirk WDJ, Terry LI. The spread of the western flower thrips Frankliniella occidentalis (Pergande) Agric Forest Entomol. 2003;5(4):301–310. doi: 10.1046/j.1461-9563.2003.00192.x. [DOI] [Google Scholar]
- 9.Murai T. Life history study of Thrips setosus. Entomologia Experimentalis et Applicata. 2001;100(2):245–251. doi: 10.1046/j.1570-7458.2001.00869.x. [DOI] [Google Scholar]
- 10.Rotenberg D, Jacobson AL, Schneweis DJ, Whitfield AE. Thrips transmission of tospoviruses. Curr Opin Virol. 2015;15:80–89. doi: 10.1016/j.coviro.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Hassani-Mehraban A, Botermans M, Verhoeven JTJ, Meekes E, Saaijer J, Peters D, Goldbach R, Kormelink R. A distinct tospovirus causing necrotic streak on Alstroemeria sp. Colombia. Arch Virol. 2010;155(3):423–428. doi: 10.1007/s00705-010-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres R, Larenas J, Fribourg C, Romero J. Pepper necrotic spot virus, a new tospovirus infecting solanaceous crops in Peru. Arch Virol. 2012;157(4):609–615. doi: 10.1007/s00705-011-1217-3. [DOI] [PubMed] [Google Scholar]
- 13.Yin Y, Zheng K, Dong J, Fang Q, Wu S, Wang L, Zhang Z. Identification of a new tospovirus causing necrotic ringspot on tomato in China. Virol J. 2014;11:213. doi: 10.1186/s12985-014-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rybicki EP. A top ten list for economically important plant viruses. Arch Virol. 2015;160(1):17–20. doi: 10.1007/s00705-014-2295-9. [DOI] [PubMed] [Google Scholar]
- 15.Scholthof K-BG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12(9):938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voller A, Bartlett A, Bidwell DE, Clark MF, Adams AN. The detection of viruses by enzyme-linked immunosorbent assay (ELISA) J Gen Virol. 1976;33(1):165–167. doi: 10.1099/0022-1317-33-1-165. [DOI] [PubMed] [Google Scholar]
- 17.Chen T-C, Lu Y-Y, Cheng Y-H, Li J-T, Yeh Y-C, Kang Y-C, Chang C-P, Huang L-H, Peng J-C, Yeh S-D. Serological relationship between Melon yellow spot virus and Watermelon silver mottle virus and differential detection of the two viruses in cucurbits. Arch Virol. 2010;155(7):1085–1095. doi: 10.1007/s00705-010-0688-y. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y-H, Dong J, Chien W-C, Zheng K, Wu K, Yeh S-D, Sun J-H, Wang Y-C, Chen T-C. Monoclonal antibodies for differentiating infections of three serological-related tospoviruses prevalent in Southwestern China. Virol J. 2016;13:72. doi: 10.1186/s12985-016-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y-H, Chen T-C, Hsu H-T, Liu F-L, Chu F-H, Chen C-C, Lin Y-Z, Yeh S-D. Serological comparison and molecular characterization for verification of calla lily chlorotic spot virus as a new tospovirus species belonging to watermelon silver mottle virus serogroup. Phytopathology. 2005;95(12):1482–1488. doi: 10.1094/PHYTO-95-1482. [DOI] [PubMed] [Google Scholar]
- 20.Mandal B, Jain RK, Krishnareddy M, Krishna Kumar NK, Ravi KS, Pappu HR. Emerging problems of tospoviruses (Bunyaviridae) and their management in the Indian Subcontinent. Plant Dis. 2012;96(4):468–479. doi: 10.1094/PDIS-06-11-0520. [DOI] [PubMed] [Google Scholar]
- 21.Chen T-C, Hsu H-T, Jain RK, Huang C-W, Lin C-H, Liu F-L, Yeh S-D. Purification and serological analyses of tospoviral nucleocapsid proteins expressed by Zucchini yellow mosaic virus vector in squash. J Virol Methods. 2005;129(2):113–124. doi: 10.1016/j.jviromet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Kuwabara K, Yokoi N, Ohki T, Tsuda S. Improved multiplex reverse transcription-polymerase chain reaction to detect and identify five tospovirus species simultaneously. J Gen Plant Pathol. 2010;76(4):273–277. doi: 10.1007/s10327-010-0246-1. [DOI] [Google Scholar]
- 23.Uga H, Tsuda S. A one-step reverse transcription-polymerase chain reaction system for the simultaneous detection and identification of multiple tospovirus infections. Phytopathology. 2005;95(2):166–171. doi: 10.1094/PHYTO-95-0166. [DOI] [PubMed] [Google Scholar]
- 24.van der Vlugt RAA, van Raaij H, de Weerdt M, Bergervoet JHW. Multiplex detection of plant pathogens through the Luminex MagPlex bead system. Methods Mol Biol. 2015;1302:283–299. doi: 10.1007/978-1-4939-2620-6_21. [DOI] [PubMed] [Google Scholar]
- 25.Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45(9):2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Brunschot SL, Bergervoet JHW, Pagendam DE, de Weerdt M, Geering ADW, Drenth A, van der Vlugt RAA. A bead-based suspension array for the multiplexed detection of begomoviruses and their whitefly vectors. J Virol Methods. 2014;198:86–94. doi: 10.1016/j.jviromet.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 27.van Brunschot SL, Bergervoet JHW, Pagendam DE, de Weerdt M, Geering ADW, Drenth A, van der Vlugt RAA. Development of a multiplexed bead-based suspension array for the detection and discrimination of pospiviroid plant pathogens. PLoS One. 2014;9(1):e84743. doi: 10.1371/journal.pone.0084743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim MS, Kim SM, Choi SH. Simultaneous detection of three lily-infecting viruses using a multiplex Luminex bead array. J Virol Methods. 2016;231:34–37. doi: 10.1016/j.jviromet.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Menzel W, Jelkmann W, Maiss E. Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. J Virol Methods. 2002;99(1–2):81–92. doi: 10.1016/S0166-0934(01)00381-0. [DOI] [PubMed] [Google Scholar]
- 30.Botermans M, van de Vossenberg BTLH, Verhoeven JTJ, Roenhorst JW, Hooftman M, Dekter R, Meekes ETM. Development and validation of a real-time RT-PCR assay for generic detection of pospiviroids. J Virol Methods. 2013;187(1):43–50. doi: 10.1016/j.jviromet.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Bald-Blume N, Bergervoet JHW, Maiss E. Development of a molecular assay for the detection of Cucumber mosaic virus and the discrimination of its subgroups I and II. J Virol Methods. 2017;243:35–43. doi: 10.1016/j.jviromet.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Bergervoet JHW, Peters J, van Beckhoven JRCM, van den Bovenkamp GW, Jacobson JW, van der Wolf JM. Multiplex microsphere immuno-detection of potato virus Y, X and PLRV. J Virol Methods. 2008;149(1):63–68. doi: 10.1016/j.jviromet.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Croft H, Malinowski T, Krizbai L, Mikec I, Kajic V, Reed C, Varga A, James D. Use of Luminex xMAP-derived Bio-Plex bead-based suspension array for specific detection of PPV W and characterization of epitopes on the coat protein of the virus. J Virol Methods. 2008;153(2):203–213. doi: 10.1016/j.jviromet.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Charlermroj R, Himananto O, Seepiban C, Kumpoosiri M, Warin N, Gajanandana O, Elliott CT, Karoonuthaisiri N. Antibody array in a multiwell plate format for the sensitive and multiplexed detection of important plant pathogens. Anal Chem. 2014;86(14):7049–7056. doi: 10.1021/ac501424k. [DOI] [PubMed] [Google Scholar]
- 35.Charoenvilaisiri S, Seepiban C, Bhunchoth A, Warin N, Luxananil P, Gajanandana O. Development of a multiplex RT-PCR-ELISA to identify four distinct species of tospovirus. J Virol Methods. 2014;202:54–63. doi: 10.1016/j.jviromet.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Hanssen IM, Lapidot M, Thomma BP. Emerging viral diseases of tomato crops. Mol Plant Microbe Interact. 2010;23(5):539–548. doi: 10.1094/MPMI-23-5-0539. [DOI] [PubMed] [Google Scholar]
- 37.Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L, Zurcher EJ (1997) Plant viruses online: descriptions and lists from the VIDE database. http://sdb.im.ac.cn/vide. Accessed 04 May 2016
- 38.Chen T-C, Li J-T, Lin Y-P, Yeh Y-C, Kang Y-C, Huang L-H, Yeh S-D. Genomic characterization of Calla lily chlorotic spot virus and design of broad-spectrum primers for detection of tospoviruses. Plant Pathol. 2012;61(1):183–194. doi: 10.1111/j.1365-3059.2011.02484.x. [DOI] [Google Scholar]
- 39.Chu F-H, Chao C-H, Chung M-H, Chen C-C, Yeh S-D. Completion of the genome sequence of watermelon silver mottle virus and utilization of degenerate primers for detecting tospoviruses in five serogroups. Phytopathology. 2001;91(4):361–368. doi: 10.1094/PHYTO.2001.91.4.361. [DOI] [PubMed] [Google Scholar]
- 40.Huang Q, Baum L, Fu WL. Simple and practical staining of DNA with GelRed in agarose gel electrophoresis. Clin Lab. 2010;56(3–4):149–152. [PubMed] [Google Scholar]


