Abstract
Since early 2006, porcine epidemic diarrhea virus (PEDV) has been reemerging in immunized swine herds. Open reading frame 3 (ORF3) is the only accessory gene in the PEDV genome. The entire ORF3 genes of 12 PEDV field strains and one vaccine strain were sequenced. The ORF3 genes of Chinese PEDV field strains (excluding CH/GSJIII/07) contain a single 672- or 675-nucleotide (nt) ORF, which encodes a 223- or 224-aa-long peptide. However, the CV777 vaccine strain and CH/GSJIII/07 contain a 276-nt ORF because of a 49-nt deletion at nt 245–293. The Chinese PEDV field strains and PEDV reference strains are divided into three groups based on the phylogenetic relationship of their ORF3 genes. Chinese PEDV field strains (excluding CH/GSJIII/07) have a close phylogenetic relationship to Korean strains and are genetically different from the PEDV vaccine strains. However, CH/GSJIII/07 has a close phylogenetic relationship to two vaccine strains, suggesting that it might have evolved from a live vaccine strain. Chinese PEDV field strains (excluding CH/GSJIII/07) can be differentiated from PEDV vaccine strains by a nested RT-PCR method.
Keywords: Porcine epidemic diarrhea virus, The phylogenetic relationship, ORF3, New genotype, Nested RT-PCR
Introduction
Porcine epidemic diarrhea virus (PEDV) is a member of Group 1a, genus Alphacoronavirus, family Coronaviridae, order Nidovirales, and is an enveloped, single-stranded, positive-sense RNA virus. The size of its genome is approximately 28 kb. Porcine epidemic diarrhea (PED), caused by PEDV, is an acute and highly contagious enteric disease that is characterized by severe enteritis, vomiting and watery diarrhea in swine. PED was firstly reported in the UK in 1971, and the virus was firstly identified during episodes of epizootic diarrhea in Belgium and the UK in 1978 [1]. This disease has subsequently been reported in many other swine-producing countries in Europe and Asia [2].
In China, transmissible gastroenteritis (TGE)-like outbreaks of acute diarrhea were first observed in Shanghai in 1973, but the causative agent of the disease was not confirmed as PEDV by fluorescent antibody test and serum neutralization test until 1984 [3]. Since 1995, bi-combined killed or attenuated vaccines have been used to against TGEV and PEDV infection in China [4, 5]. However, since early 2006, PEDV has been emerging in immunized swine herds in spite of using the current vaccine strategy. The damage caused by PEDV infection has been continuous and serious, and the diarrhea caused by PEDV cannot be differentiated clinically from acute TGE. Sometimes, the diarrhea caused by PEDV and TGEV coinfection occurs in the field.
A variety of genes encoding accessory proteins are interspersed between the structural genes. Their number and location vary among coronavirus genomes. These genes are called accessory genes. Whereas the genes encoding the structural proteins have been thoroughly investigated for most coronaviruses, little is known about the accessory genes. Reverse genetic analyses of mouse hepatits virus (MHV) and feline infectious peritonitis virus (FIPV) have suggested that these genes are not required for virus replication. Moreover, deletion of MHV and FIPV accessory genes results in attenuation in their respective hosts, indicating that the accessory genes represent pathogenicity factors [6–8]. It has been shown that virulence of TGEV and PEDV can be reduced by altering the ORF3 gene through cell-culture adaptation [9, 10], and ORF3 has been suggested to be an important determinant for the virulence of these viruses. ORF3 is the only accessory gene in the PEDV genome. Differentiation of ORF3 genes between the highly Vero-cell-adapted viruses and field virus could be a marker of adaptation to cell culture and a valuable tool for molecular epidemiologic studies of PEDV infections [10].
In order to control and prevent PEDV infection, it is necessary for us to further investigate the molecular epidemiology of PEDV field strains in China. The purpose of this study is to investigate the molecular epidemiology and diversity among Chinese PEDV field strains based on sequence analysis and examination of the phylogenetic relationship of ORF3 genes and to establish a method that can differentiate PEDV field strains from PEDV vaccine strains.
Materials and methods
Porcine fecal samples were taken from piglets with watery diarrhea and dehydration at 11 different swine-raising farms in 8 provinces in China (Gansu, Heilongjiang, Henan, Hunan, Inner Mongolia, Jiangsu, Jilin and Shanghai) where the bi-combined killed or attenuated vaccines against TGEV and PEDV infection were used from January 2006 to August 2009. These fecal samples had been confirmed positive for PEDV by RT-PCR or by using an Anigen Rapid PED Ag Test Kit (Animal Genetics Inc., Korea) [11]. PEDV-positive fecal samples were diluted with phosphate-buffered saline (PBS; 0.1 M, pH 7.2) to make 10% (V/V) suspensions. The suspensions were vortexed and clarified by centrifugation for 10 min at 5,000 rpm. The supernatants were collected for RT-PCR or nested RT-PCR.
Two pairs of primers (Table 1) for nested RT-PCR were designed and synthesized according to the sequence of PEDV CV777 strain (GenBank accession number AF353511). ORF3U1 and ORF3L1 were used for the amplification of the full ORF3 gene of PEDV. The size of the expected product was 774 bp. ORF3U2 and ORF3L2 were used to differentiate Chinese field PEDV strains from the CV777 vaccine strain. The size of the expected product was 149 bp.
Table 1.
Sequence and location of the oligonucleotides used for nested RT-PCR
| Primers | Sequence | Size (bp)a | Location in genomeb |
|---|---|---|---|
| ORF3U1 | 5′-CCTAGACTTCAACCTTACGA-3′ | 774 | 24,742–24,761 |
| ORF3L1 | 5′-CAGGAAAAAGAGTACGAAAA-3′ | 25,515–25,496 | |
| ORF3U2 | 5′-ACTGTGGTGCACTTTTAGAT-3′ | 149 | 25,051–25,070 |
| ORF3L2 | 5′-AGCTGCTTTACCATTGAGAA-3′ | 25,199–25,180 |
aPredicted from the sequence
bIn relation to the genome of PEDV CV777 strain (AF353511)
Viral RNA was extracted from the supernatants or the live vaccine using TRIzol Reagent (Invitrogen Corp., Carlsbad, USA) according to the manufacturer’s instructions. The RNA pellet was dissolved in diethylpyrocarbonate (DEPC)-treated deionized water, and the first-strand complementary DNA (cDNA) was synthesized using ORF3L1 as the reverse transcription primer as described previously [11].
PCR was carried out in a two-step reaction, first with a pair of primers flanking the region to be amplified (ORF3U1 and ORF3L1), and then using a pair of primers within the amplified sequence (ORF3U2 and ORF3L2) in a total volume of 25 µl. In detail, 1 µl cDNA was mixed with a reaction mixture containing 2.5 µl 10× Ex Taq buffer (TaKaRa, Dalian, China), 2 μl 2.5 mM dNTPs (TaKaRa), 0.5 µl each specific primer (10 pmol), 0.25 µl Ex Taq DNA polymerase (TaKaRa) and 18.25 μl sterile deionized water. The first round of amplification was performed under the following reaction conditions: pre-denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 60 s, annealing at 50°C for 60 s, extension at 72°C for 60 s, and a final extension at 72°C for 10 min. For the second round of amplification, a 100-fold dilution of the first PCR product in distilled water was used as a template under the following reaction conditions: pre-denaturation at 94°C for 5 min followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min.
PCR products were excised from 1.0% agarose gels and purified using an AxyPrep™ DNA Gel Extraction Kit (Axygen Scientific, Inc., USA) and then cloned into a T-tailed vector, pMD18-T and introduced into competent JM109 cells (TaKaRa) by transformation. Three recombinant DNA clones of each PEDV strains were sequenced by Beijing Genomics Institute (China). The nucleotide sequences of 13 PEDV strains (including CV777 vaccine strain) were edited, analyzed with EditSeq software and aligned with reference sequences using MegAlign software (version 7.1.0, DNASTAR Inc., USA).
The sequences of PEDV strains described in the present study have been deposited in the GenBank database. The accession numbers are as follows: CH/HLJH/06 (GU372732), CH/S (GU372733), CH/JL/08 (GU372734), CH/HLJM/07 (GU372735), CH/HNHJ/08 (GU372736), CH/GSJI/07 (GU372737), CH/HNCH/06 (GU372738), CH/IMT/06 (GU372739), CH/SHH/06 (GU372740), CH/JL/09 (GU372741), CH/GSJII/07 (GU372742), CH/GSJIII/07 (GU372743) and CV777 vaccine strain (GU372744). The accession numbers for reference PEDV strains are as follows: CV777 (AF353511), Br1/87 (Z24733), LZC (EF185992), DR13 (EU054929), DR13 vaccine strain (EU054930) and Chinju99 (EU792474).
The sequences of ORF3 genes were also aligned with ClustalX v1.83, and phylogenetic analysis was conducted by the minimum-evolution method, Kimura distances and a bootstrap of 1,000 replicates, using the MEGA program [12].
Results
The approximately 774-bp fragments containing the full ORF3 gene were amplified by the first pair of primers, ORF3U1/ORF3L1, from all of the field strains and the CV777 vaccine strain (Fig. 1a). The 149-bp products were amplified by the second pair of primers, ORF3U2/ORF3L2, from the field strains (excluding CH/GSJIII/07). However, no PCR products were amplified by ORF3U2/ORF3L2 from the CH/GSJIII/07 and CV777 vaccine strain (Fig. 1b).
Fig. 1.

Nested RT-PCR on the PEDV strains (including CV777 vaccine strain). a The first PCR products for the 11 field PEDV strains and the CV777 vaccine strain. b The second PCR products for the 11 field PEDV strains and CV777 vaccine strain. From left to right: A CH/HLJH/06, B CH/HNCH/06, C CH/IMT/06, D CH/SHH/06, E CH/HLJM/07, F CH/GSJI/07, G CH/GSJII/07, H CH/HNHJ/08, I CH/JL/08, J CH/JL/09, K CH/GSJIII/07, L CV777 vaccine strain, M negative control, N DNA marker (DL2,000)
The ORF3 genes of Chinese PEDV field strains (excluding CH/GSJII/07 and CH/GSJIII/07) contain a single ORF, which consists of 675 nucleotides and encodes a 224-aa-long peptide. CH/GSJII/07 has a 3-nt deletion at nt 416–418 (Fig. 2) and contains a single 672-nt ORF encoding a 223-aa-long peptide. However, the ORF3 genes of the CV777 vaccine strain and CH/GSJIII/07 have a 49-nt deletion at nt 245–293 compared with the parent CV777 and other Chinese field strains. In addition to the large deletion region, CH/GSJIII/07 also has 3 nt deletions at nt 417–419 (Fig. 2).
Fig. 2.
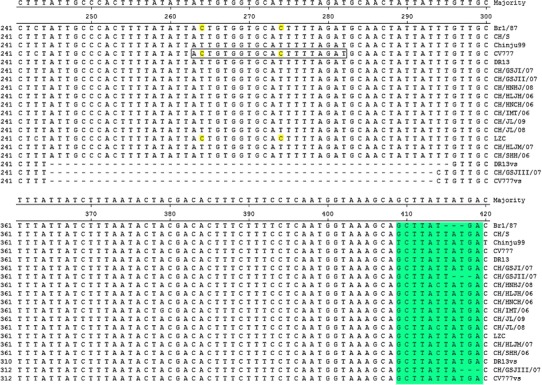
Comparison of partial nucleotide sequences of ORF3 genes of Chinese PEDV strains and PEDV reference strains. The dashes indicate deleted nucleotides. The oligonucleotide in the box is the primer ORF3U2, and the nucleotides (nt 264, 274) are identical in Br1/87, CV777 and LZC, but different from those in other PEDV strains. vs vaccine strain. The region (nt 409–419) is the previously reported variable region II
Nucleotide and deduced amino acid sequence homology results are described in Table 2. The recent Chinese field strains (excluding CH/GSJIII/07) have 97.6–100% DNA sequence identity to each other. They share more than 97.2% DNA sequence identity with Korean field strains (Chinju99, DR13). However, they have 96.6–97.6% DNA sequence identity to CV777 vaccine strain, less than 95.9% DNA sequence homology to Chinese stain LZC and less than 97.2% DNA sequence identity to the European field strains (CV777, Br1/87). The deduced amino acid sequences of the recent Chinese field strains (excluding CH/GSJIII/07) have 96.4–100% homology to each other and more than 95.2% to those of Korean strains (Chinju99, DR13 and attenuated DR13). They have less than 94.6% homology to the deduced amino acid sequences of two European strains (CV777, Br1/87) and one Chinese strain (LZC). CH/GSJIII/07 has 99.0% DNA sequence homology with CV777 vaccine strain and 100% homology to the deduced amino acid sequence of the CV777 vaccine strain.
Table 2.
Comparison of the nucleotide and deduced amino acid sequences of ORF3 genes of Chinese PEDV strains and PEDV reference strains
| Strain/isolate | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Br1/87 | *** | 95.8 | 96.4 | 96.1 | 96.1 | 95.8 | 95.8 | 96.3 | 95.7 | 96.3 | 95.4 | 96.0 | 96.3 | 96.1 | 99.0 | 95.2 | 96.1 | 95.7 | 97.6 |
| 2 CH/GSJI/07 | 94.2 | *** | 98.4 | 96.3 | 99.7 | 99.3 | 99.0 | 99.1 | 99.1 | 99.1 | 98.8 | 98.4 | 99.1 | 98.2 | 96.9 | 97.3 | 98.7 | 97.4 | 95.6 |
| 3 CH/GSJII/07 | 94.6 | 98.2 | *** | 98.1 | 98.7 | 98.1 | 97.8 | 97.9 | 97.9 | 97.9 | 97.6 | 97.2 | 97.9 | 97.2 | 96.1 | 96.6 | 97.6 | 96.8 | 94.8 |
| 4 CH/GSJIII/07 | 86.8 | 87.9 | 87.9 | *** | 96.6 | 96.3 | 96.3 | 96.5 | 96.1 | 96.5 | 95.8 | 96.0 | 96.5 | 96.0 | 95.8 | 99.0 | 96.1 | 97.9 | 94.4 |
| 5 CH/HLJH/06 | 94.2 | 100.0 | 98.2 | 87.9 | *** | 99.4 | 99.1 | 99.3 | 99.3 | 99.3 | 99.0 | 98.5 | 99.3 | 98.5 | 97.2 | 97.6 | 99.0 | 97.8 | 95.9 |
| 6 CH/HLJM/07 | 93.7 | 98.7 | 96.9 | 89.0 | 98.7 | *** | 99.4 | 99.6 | 99.9 | 99.6 | 99.6 | 98.8 | 99.6 | 98.5 | 96.9 | 97.3 | 98.7 | 97.4 | 95.6 |
| 7 CH/HNCH/06 | 94.2 | 99.1 | 97.3 | 89.0 | 99.1 | 99.6 | *** | 99.3 | 99.3 | 99.3 | 99.0 | 98.8 | 99.3 | 98.5 | 96.9 | 97.3 | 98.4 | 97.4 | 95.6 |
| 8 CH/HNHJ/08 | 94.2 | 99.1 | 97.3 | 89.0 | 99.1 | 99.6 | 100.0 | *** | 99.4 | 100.0 | 99.1 | 98.7 | 100.0 | 98.7 | 97.2 | 97.4 | 98.5 | 97.6 | 95.9 |
| 9 CH/IMT/06 | 93.3 | 98.2 | 96.4 | 89.0 | 98.2 | 99.6 | 99.1 | 99.1 | *** | 99.4 | 99.4 | 98.7 | 99.4 | 98.4 | 96.7 | 97.1 | 98.5 | 97.3 | 95.4 |
| 10 CH/JL/08 | 94.2 | 99.1 | 97.3 | 89.0 | 99.1 | 99.6 | 100.0 | 100.0 | 99.1 | *** | 99.1 | 98.7 | 100.0 | 98.7 | 97.2 | 97.4 | 98.5 | 97.6 | 95.9 |
| 11 CH/JL/09 | 93.7 | 98.7 | 96.9 | 89.0 | 98.7 | 100.0 | 99.6 | 99.6 | 99.6 | 99.6 | *** | 98.4 | 99.1 | 98.1 | 96.4 | 96.8 | 98.2 | 97.0 | 95.1 |
| 12 CH/S | 94.2 | 99.1 | 97.3 | 89.0 | 99.1 | 99.6 | 100.0 | 100.0 | 99.1 | 100.0 | 99.6 | *** | 98.7 | 98.2 | 97.0 | 97.0 | 98.7 | 97.1 | 95.7 |
| 13 CH/SHH/06 | 94.2 | 99.1 | 97.3 | 89.0 | 99.1 | 99.6 | 100.0 | 100.0 | 99.1 | 100.0 | 99.6 | 100.0 | *** | 98.7 | 97.2 | 97.4 | 98.5 | 97.6 | 95.9 |
| 14 Chinju99 | 93.7 | 98.7 | 96.9 | 89.0 | 98.7 | 98.2 | 98.7 | 98.7 | 97.8 | 98.7 | 98.2 | 98.7 | 98.7 | *** | 97.2 | 97.0 | 98.1 | 97.1 | 95.9 |
| 15 CV777 vs | 86.8 | 87.9 | 87.9 | 100.0 | 87.9 | 89.0 | 89.0 | 89.0 | 89.0 | 89.0 | 89.0 | 89.0 | 89.0 | 89.0 | *** | 96.8 | 97.2 | 97.3 | 98.7 |
| 16 CV777 | 97.8 | 96.4 | 94.6 | 86.8 | 96.4 | 96.0 | 96.4 | 96.4 | 95.5 | 96.4 | 96.0 | 96.4 | 96.4 | 96.0 | 86.8 | *** | 97.1 | 99.2 | 95.4 |
| 17 DR13 | 94.6 | 99.6 | 97.8 | 89.0 | 99.6 | 99.1 | 99.6 | 99.6 | 98.7 | 99.6 | 99.1 | 99.6 | 99.6 | 99.1 | 89.0 | 96.9 | *** | 97.3 | 95.9 |
| 18 DR13 vs | 93.2 | 97.6 | 95.2 | 89.0 | 97.6 | 97.1 | 97.6 | 97.6 | 96.6 | 97.6 | 97.1 | 97.6 | 97.6 | 97.1 | 89.0 | 96.1 | 98.1 | *** | 95.8 |
| 19 LZC | 96.0 | 94.6 | 92.8 | 85.7 | 94.6 | 94.2 | 94.6 | 94.6 | 93.8 | 94.6 | 94.2 | 94.6 | 94.6 | 94.2 | 85.7 | 98.2 | 95.1 | 94.2 | *** |
Nucleotide identity (%) in upper triangle
Deduced amino acid identity (%) in lower triangle
Based on the phylogenetic analysis of the ORF3 genes, all of the PEDV strains are divided into three groups (Fig. 3). Group I comprises 11 Chinese field strains and 2 Korean strains (DR13, Chinju99). Group II consists of one Chinese field strain (CH/GSJIII/07) and two vaccine strains (attenuated CV777 and attenuated DR13). Group III is composed of one Chinese strain (LZC) and two European strains (CV777 and Br1/87).
Fig. 3.
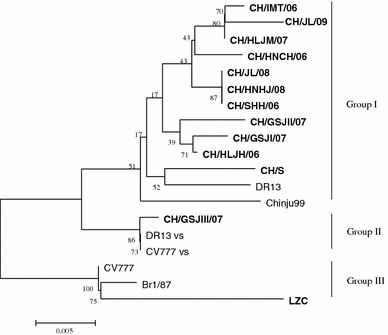
Phylogenetic analysis of the nucleotide sequences of ORF3 genes of Chinese PEDV strains and PEDV reference strains. The tree was constructed based on the minimum-evolution method using MEGA 4 software. Numbers above branches indicate bootstrap values calculated from 1,000 bootstrap replicates. The scale bar indicates the branch lengths for 0.5% nucleotide differences. The Chinese PEDV strains are in bold
Discussion
In this study, the complete sequences of the ORF3 genes of Chinese PEDV field strains and the CV777 vaccine strain have been determined. There are no insertions or deletions in the ORF3 genes of the Chinese PEDV field strains (excluding CH/GSJII/07 and CH/GSJIII/07)—only point mutations. All of the ORF3 genes of the Chinese PEDV field strains and the CV777 vaccine strain have a sequence (CTAGAC) of 46 nucleotides upstream of the initiator ATG, as already recognized in a previous study [13]. This sequence is a hexameric motif common to coronaviruses and is similar to the hexameric motifs XUA(A/G)AC found adjacent to other PEDV ORFs. These hexameric motifs have been proposed to be a start site for transcription of the subgenomic mRNAs [14].
A previous study has indicated that there are three variable regions (I, II and III) in the ORF3 gene [13]. In our study, variable regions I and III are not observed in all the ORF3 genes of Chinese field strains and CV777 vaccine strain. However, variable region II is observed in the ORF3 genes of CH/GSJII/07 and CH/GSJIII/07. Both CH/GSJII/07 and CH/GSJIII/07 have a 3-nt deletion in variable region II (Fig. 2). Both the CV777 vaccine strain and CH/GSJIII/07 have a 49-nt deletion at nt 245–293. The large deletion is similar to the deletion in the ORF3 gene of attenuated DR13, which has a 51-nt deletion at nt 245–295 (Fig. 2). Although they have 51-nt deletions in their ORF3 genes, attenuated-type PEDV strains, including attenuated DR13, KPED-9 and P-5V, contain a single 624-base ORF encoding a 224-aa-long peptide [15]. However, because of the large deletion, the CV777 vaccine strain and CH/GSJIII/07 contain a truncated ORF, which consists of 276 nt and encodes a 91-aa-long peptide.
Previous studies revealed that the highly cell-culture-adapted PEDV, attenuated DR13, could be differentiated from wild-type PEDV strains by both RT-PCR and restriction fragment length polymorphism (RFLP), which used sequence variations in the ORF3 gene of the highly cell-culture-adapted PEDV [10, 15]. Because detection of PEDV in feces might be affected by the reliability and sensitivity of the technique [16], we have established a nested RT-PCR method using the deletion in the ORF3 gene of the CV777 vaccine strain. Because the sense primer ORF3U2 is located in the region that corresponds to the large deletions in the ORF3 genes of PEDV vaccine strains (Fig. 2), the 149-bp PCR products could not be amplified by ORF3U2/ORF3L2 from CV777 vaccine strain when using the products amplified by ORF3U1/ORF3L1 as the templates. In this study, the 149-bp fragments of the CV777 vaccine strain and CH/GSJIII/07 were not amplified, while the corresponding fragments of other Chinese PEDV field strains were amplified by the nested RT-PCR (Fig. 1).
In order to trace the origin and evolution of PEDV field strains in China, a phylogenetic tree of the Chinese PEDV field strains and PEDV reference strains was constructed based on the nucleotide sequences of the ORF3 genes. The phylogenetic relationship of the ORF3 genes indicates that Chinese PEDV field strains (excluding CH/GSJIII/07) differ genetically from European field strains (CV777 and Br1/87) and have a close phylogenetic relationship to Korean field strains (DR13 and Chinju99). The Chinese PEDV field strains (excluding CH/GSJIII/07) are genetically different from the CV777 vaccine strain, which is used to prevent PEDV infection in China at present. There is a new genotype of PEDV prevailing in China that differs from the genotype of the vaccine strains. PEDV shedding may be variable, depending on the sensitivity of the detection tool. Moreover, shedding may be influenced by the viral strain employed. It has been reported that virus shedding in piglets inoculated with cell-adapted DR13 lasted for 7 or 8 days [10, 17], while in sows, it persisted for 3 days [17]. CH/GSJIII/07 has the highest DNA sequence identity and a close phylogenetic relationship to the CV777 vaccine strain. It may have evolved from the vaccine strain under the immune pressure. However, further work will be required to determine whether this field strain is a variant of the CV777 vaccine strain.
By accurate analysis of ORF3 genes, we can understand the molecular epidemiology of PEDV field strains in our country. Differentiation of ORF3 genes between highly Vero-cell-adapted PEDV and PEDV field strains can help us to choose an appropriate PEDV field strain as a vaccine candidate and prevent outbreaks of PEDV-induced diarrhea more effectively. Moreover, the complete nucleotide sequences of the ORF3 genes of the CV777 vaccine strain and the 12 field samples will now form the basis for further functional exploration of both wild- and attenuated-type PEDV strains in China.
Acknowledgments
This study was supported by National Key Laboratory of Veterinary Biotechnology (NKLVBP200805), National Natural Science Foundation of China (30901081), and National Key Technology Research and Development Program (2006BAD06A07). We also thank Dr. Tongqing An for providing ClustalX v1.83 and MEGA 4 softwares and giving advice.
References
- 1.Pensaert MB, de Bouck P. A new coronavirus-like particles associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pensaert MB, Yeo SG. Porcine epidemic diarrhea. In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ, editors. Disease of Swine. 9. Ames: Blackwell; 2006. pp. 367–372. [Google Scholar]
- 3.Xuan H, Xing D, Wang D, Zhu W, Zhao F, Gong H. Study on the culture of porcine epidemic diarrhea virus adapted to fetal porcine intestine primary cell monolayer. Chin J Vet Sci. 1984;4(3):202–208. [Google Scholar]
- 4.Ma S, Wang M, Feng L, Li W. Study on Bi-combined killed vaccine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Chin J Prev Vet Med. 1995;17(6):23–27. [Google Scholar]
- 5.Tong Y, Feng L, Li W, Zhu Y, Wang M, Ma S. Development of Bi-combined attenuated vaccine against transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Chin J Prev Vet Med. 1999;21(6):406–410. [Google Scholar]
- 6.de Haan CA, Masters PS, Shen X, Weiss S, Rottier PJ. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrewegh AA, Vennema H, Horzinek MC, Rottier PJ, de Groot RJ. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212:622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haijema BJ, Volders H, Rottier PJ. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woods RD. Efficacy of a transmissible gastroenteritis coronavirus with an altered ORF 3 gene. Can J Vet Res. 2001;65:28–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Song DS, Yang JS, Oh JS, Han JH, Park BK. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21:1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JF, Sun DB, Wang CB, Shi HY, Cui XC, Liu SW, Qiu HJ, Feng L. Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes. 2008;36:355–364. doi: 10.1007/s11262-007-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Duarte M, Tobler K, Bridgen A, Rasschaert D, Ackermann M, Laude H. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology. 1994;198:466–476. doi: 10.1006/viro.1994.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte M, Gelfi J, Lambert P, Rasschaert D, Laude H. Genome organization of porcine epidemic diarrhoea virus. Adv Exp Med Biol. 1993;342:55–60. doi: 10.1007/978-1-4615-2996-5_9. [DOI] [PubMed] [Google Scholar]
- 15.Park SJ, Moon HJ, Luo Y, Kim HK, Kim EM, Yang JS, Song DS, Kang BK, Lee CS, Park BK. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guscetti F, Bernasconi C, Tobler K, Van Reeth K, Pospischil A, Ackerman M. Immunohistochemical detection of porcine epidemic diarrhea virus compared to other method. Clin Diagn Lab Immunol. 1998;5:412–414. doi: 10.1128/cdli.5.3.412-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song DS, Oh JS, Kang BK, Yang JS, Song JY, Moon HJ, Kim TY, Yoo HS, Jang YS, Park BK. Fecal shedding of a highly cell adapted porcine epidemic diarrhea virus after oral inoculation in pigs. J Swine Health Prod. 2005;13(5):269–272. [Google Scholar]


