Abstract
Avian infectious bronchitis virus (IBV) is a member of the family Coronaviridae. A binding domain that mediates the attachment of the virus to its receptor has been identified in the S1 protein of prototype IBV strain M41. In this study, we identified this binding domain in a different strain, as well as the cellular proteins that interact with it. First, we expressed the S1N proteins (residues 19-270) of M41 and another isolate, SCZJ3, and compared the binding capacities of recombinant S1N-M41 and S1N-SCZJ3 to host tissues. Protein histochemistry showed that both S1N-M41 and S1N-SCZJ3 could bind to lung and kidney, and that recombinant S1N-SCZJ3 displayed a distinctive staining pattern in the proventriculus. Recombinant S1N-SCZJ3 was then employed to purify binding-associated proteins in lung, kidney, and proventriculus. Using an affinity chromatography assay, two common bands of about 60 kDa and 70 kDa were obtained from the total tissue proteins. These protein bands were identified by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) as protein disulfide isomerase (PDI) and heat shock protein 70 (HSP70). Finally, infection of chicken embryo kidney (CEK) cells by SCZJ3 was found to be inhibited by anti-HSP70 but not anti-PDI polyclonal antibody. These data indicate that HSP70 is part of the receptor complex of IBV and might help to understand the mechanism of S-mediated cell entry of IBV.
Electronic supplementary material
The online version of this article (doi:10.1007/s00705-017-3280-x) contains supplementary material, which is available to authorized users.
Keywords: Sialic Acid, Protein Disulfide Isomerase, Infectious Bronchitis Virus, Japanese Encephalitis Virus, Infectious Bronchitis Virus Strain
Introduction
Infectious bronchitis virus (IBV) is a member of the genus Gammacoronavirus. It is an enveloped virus with a positive-strand RNA genome, which is highly variable. IBV variants of different genotypes have been found worldwide. Mutations have been mainly found in the S1 protein that can result in poor cross-protection between genotypes. The virus can replicate in many tissues [1] and causes various pathological changes, including respiratory symptoms and lesions of the kidney and proventriculus [2].
The infection is initiated by the binding of the spike (S) protein to host tissues [3], which has been shown to be a determinant of tropism and pathogenicity [4]. A functional region has been identified on the S1 subunit of several coronavirus, called the receptor-binding domain (RBD) [5]. Certain residues in the RBD are critical for binding activity, and single mutations may abolish or change the binding activity of the virus [6]. In the case of IBV, the N-terminal portion of S1 (residues 19-272) was mapped as the binding domain of the Mass type strain M41 [7]. However, the S1 subunit is highly variable, and mutations have been found in S1 of different genotypes [8]. These mutations may influence the binding activity of RBD to host tissues. Thus, we attempted to identify the receptor-binding domain of a different genotype.
Here, we have identified the binding domain of a prevalent strain SCZJ3, which has the characteristics of the QX-like genotype and shows only 75.9% amino acid sequence identity to M41 in the S1N region (residues 19-270) [9]. In vivo, M41 is a respiratory IBV strain that mainly affects the upper respiratory tract. QX-like strains have been reported to be associated with distinct lesions of the proventriculus and kidney. M41 was first isolated in 1996 and is now becoming epidemic in China [10]. To investigate differences in the pathogenicity of QX-like and Mass-like strains [11], the binding of S1N-SCZJ3 and S1N-M41 to lung, kidney and proventriculus were analyzed.
The virus-cell interaction of IBV is complicated; the binding of virus to host cells may initiate a chain of molecular interactions that enable viral entry. Sialic acid and heparan sulfate have been reported to be involved in IBV infection. Removal of sialic acid can abolish the binding of S1 to respiratory tissues [12, 13]. However, sialic acid is present on cells that are not sensitive to IBV, indicating that sialic acid is not the only molecule that binds to IBV [4]. Heparan sulfate was identified as a binding molecule only for the cell-adapted strain Beaudette, with the binding sites located within the S2 subunit (residues 686-691) [14]. Up to now, there has been little study of the protein involved in the binding process of IBV. In studies of SASR and 229E, RBDs showed specific binding to host receptors and showed higher binding activity than full-length S1 [15, 16]. This binding activity has been utilized to define the receptor of virus [17, 18]. In the present study, an affinity chromatography assay was used to isolate proteins that interacted with S1N-SCZJ3 [19]. Using an infection inhibition assay, we found that HSP70 is part of the receptor complex of IBV.
Materials and methods
Virus and cells
Infectious bronchitis virus (IBV) strains SCZJ3 (JF951370.1) and M41 (DQ834384.1) were kept in our laboratory. Spodoptera frugiperda (Sf9) cells were maintained in Sf-900™ SFM II medium (Gibco). Chicken embryo kidney (CEK) cells were generated from fifteen-day-old chicken embryos and maintained in DMEM with high glucose supplemented with 10% fetal calf serum (FCS).
Protein expression and purification
Viral RNA was extracted from allantoic fluid and reverse transcribed using a PrimerScript™ RT Reagent Kit (Takara). The sequences encoding S1N-SCZJ3 and S1N-M41 (nucleotides 55 to 810) were amplified and cloned into the Bac-to-Bac baculovirus expression system according to the manufacturer’s instructions [20].
Sf9 cell monolayers were infected with baculoviral stock at an MOI of 5 and incubated at 27 °C for 72 h before harvesting. Cells were frozen and thawed and then suspended in lysis buffer (10 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl, pH 8.0). The lysates were then centrifuged at 10,000 g for 10 min at 4 °C, and the supernatants were purified using an Ni-NTA column (Ni Sepharose™, GE Healthcare).
The purified proteins (S1N-SCZJ3 and S1N-M41) and PNGase F (NEB)-treated proteins were separated by 10% SDS-PAGE and detected using polyclonal IBV-M41 antiserum (diluted 1:100 in PBS, China Institute of Veterinary Drug Control). Western blot analysis was performed as described [21].
Immunohistochemistry
Specific-pathogen-free (SPF) eggs were purchased from Merial-Beijing and incubated in a 37 °C humidity chamber. Tissues were taken from 5-week-old chickens and fixed with 10% formalin. Protein histochemistry was performed as described [13]. S1N-SCZJ3, S1N-M41 (30 μg/ml) and PBS as a control were applied to slides and incubated at 37 °C for 2h. Binding was then detected using a monoclonal antibody against the His-tag (HRP-conjugated, diluted 1:100 with PBS) at 37 °C for 1 h.
Preparation of total proteins and affinity chromatography
Lung, kidney and proventriculus tissues were taken from 3-day-old SPF chickens and frozen with liquid nitrogen. Tissues were ground and suspended in phosphate buffer (50 mg, 1 ml) containing 1% Triton X-100 and PMSF. The lysates were subjected to ultrasonic treatment and centrifuged at 10,000 g for 10 min at 4 °C. The concentrations of the supernatants were determined using a NanoDrop 2000 spectrophotometer.
Ni-NTA resin coupled with S1N-SCZJ3 or Ni-NTA resin alone as control (1 ml) was incubated with total tissue proteins (1 ml) and rotated end over end for 4 h at 16 °C. The tube was then centrifuged at 3000 g for 2 minutes. Finally, the resins were washed four times with wash buffer (5 ml) with increasing concentrations of imidazole (20, 50, 100, 250 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl, pH 8.0), followed by 10 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 100 mM EDTA, pH 8.0). The eluates were separated by 10% SDS-PAGE, and the bands were analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).
Binding blocking assay
SCZJ3 virus (103 EID50) diluted in PBS was pretreated with increasing concentrations (22, 45, 90, 180 μg/ml) of chromatography eluates in equal volumes and incubated at 37 °C for 1 h. Chicken embryo kidney cells cultured in 6-well plates were infected with 1-ml incubation at 37 °C for 1 h, and cells were harvested after three washes with PBS. The virus loads were determined by real-time PCR as described [1].
Infection inhibition assay
Chicken embryo kidney cells were cultured in 24-well plates, and the cells were incubated with increasing concentrations of anti-PDI or anti-HSP70 polyclonal antibody for 1 h at 37 °C, or PBS as a control. The cells were then infected with SCZJ3 virus (103 EID50, 200 μl) diluted in PBS for 1 h at 37 °C. Cells were harvested after three washes with PBS. The virus loads were determined by real-time PCR.
Results
Expression, purification and histochemistry of S1N-SCZJ3 and S1N-M41
SCZJ3 and M41 are indeed two separate IBV strains in terms of both genotype and pathogenicity. The predicted amino acid sequences of the N-terminal peptides of S1 (S1N, aa 19-270) were aligned by the ClustalW method, and the genetic distance between the S1N-SCZJ3 S1N and M41 peptides was 0.315. Four main variable regions were found at aa positions 20-26, 53-80, 117-122, and 129-136, containing the amino acids critical for M41 (N38T, H43Q, P63S, T69V) (Fig. 1A). We then expressed S1N (residues 19-270) of SCZJ3 and M41 in a baculovirus-insect system. As shown in Fig. 1B, both proteins were highly glycosylated and could be detected by polyclonal IBV-M41 antiserum before and after treatment with PNGase F, indicating that S1N-SCZJ3 and S1N-M41 were expressed successfully.
Fig. 1.
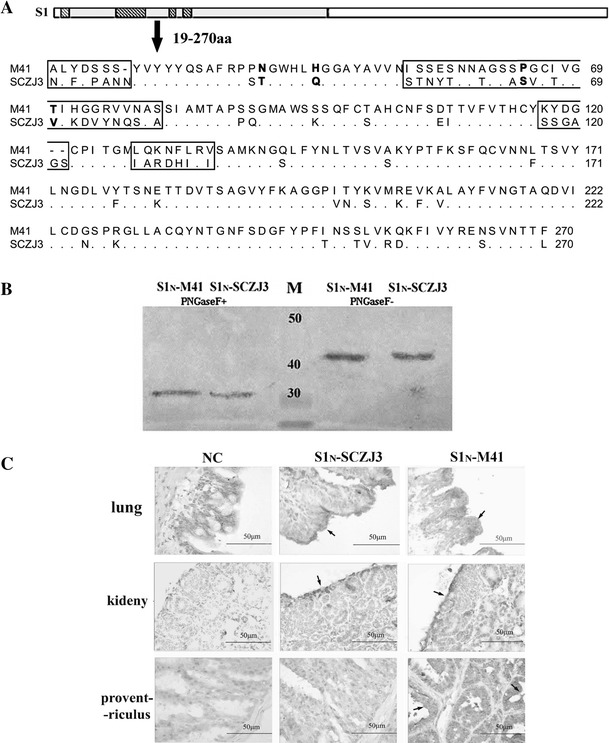
(A) Amino acid sequence alignment of S1N (residues 19–270) of M41 and SCZJ3. Four main divergent regions are located at aa 20-26, 53-80, 117-122, 129-136. The critical amino acids for M41 (N38T, H43Q, P63S, T69V) are shown in bold. (B) Western blot. S1N-SCZJ3 and S1N-M41 were purified using an Ni-NTA column and analyzed by Western blot. The samples were both recognized by polyclonal IBV-M41 antiserum before and after treatment with PNGase F. (C) Histochemistry was performed by incubating S1N-M41 and S1N-SCZJ3 with lung, kidney and proventriculus samples of PBS as control. Binding to peribronchial epithelial cells in lung tissue and to the renal tubular epithelial cells in kidney were detected. Distinctive binding of S1N-SCZJ3 to mucous membrane epithelial cells was detected in the proventriculus
To compare the biological activities of S1N-SCZJ3 and S1N-M41, we examined their ability to bind to lung, kidney and proventriculus. Binding to peribronchial epithelial cells in the lung and renal tubular epithelial cells in the kidney was detected, indicating that the S1N region of both M41 and SCZJ3 could play a role in receptor binding. In addition, S1N-SCZJ3 showed distinct binding to proventriculus mucous membrane epithelial cells (Fig 1C), which may correlate the tissue tropism of SCZJ3 [10].
Isolation of receptor-associated proteins by affinity chromatography and LC-MS/MS
Next, we decided to investigate the binding-associated proteins in lung, kidney and proventriculus. Due to the lack of cultured cell lines for IBV, we attempted to isolate proteins that interact with S1N-SCZJ3 in tissue lysates, which may increase the abundance of the obtained proteins. Tissue lysates were incubated with Ni-NTA resins containing S1N-SCZJ3 as ligand, and after extensive washing, chromatography eluates were analyzed by 10% SDS-PAGE. As shown in Fig. 2A, two main common bands were detected (60 kDa and 70 kDa), and two distinctive bands were detected in proventriculus tissue (27 kDa and 45 kDa). A similar result was obtained using a virus overlay protein binding assay (VOPBA), demonstrating that the molecules isolated using recombinant S1N-SCZJ3 are specific (Supplementary Figure 1). In tissue lysates incubated with Ni-NTA resin alone, the nonspecific bands were weak and diverse under the same elution conditions (Supplementary Figure 2).
Fig. 2.
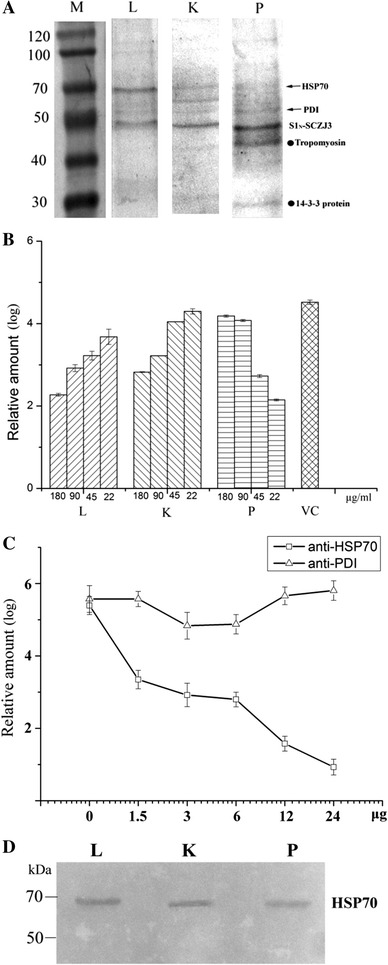
(A) Isolation of S1N-SCZJ3 binding-associated proteins by affinity chromatography. Total proteins of lung (L), kidney (K), and proventriculus (P) were incubated with S1N-SCZJ3-coupled Ni-NTA resins. The chromatography eluates were analyzed by 10% SDS-PAGE. Four bands were obtained (as indicated) and identified as 14-3-3 protein (27 kDa), tropomyosin (45 kDa), protein-disulfide isomerase (60 kDa), and heat shock protein 70 (70 kDa) by LC-MS/MS. (B) Blocking of binding by chromatography eluates. SCZJ3 (103 EID50) was pre-incubated with affinity chromatography eluates of lung (L), kidney (K), proventriculus (P) and PBS as virus control (VC). Subsequently, CEK cells were infected with 1 ml of these preparations and the viral loads were determined by real-time PCR. (C) Inhibition of infection by anti-HSP70 and anti-PDI antibodies. CEK cells were pretreated with increasing concentrations of anti-HSP70 and anti-PDI antibodies for 1 h at 37 °C. Cells were infected with SCZJ3 (103 EID50, 200 μl), and the viral loads were determined by real-time PCR. (D) Affinity chromatography eluates were incubated with anti-HSP70 antibody
The bands were then analyzed by trypsin digestion and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), followed by Mascot search analysis. The proteins were identified as 14-3-3 protein (27 kDa), tropomyosin (45 kDa), protein disulfide isomerase (PDI) (60 kDa) and heat shock protein 70 (70 kDa) (Supplementary Figure 3).
Identification of HSP70 as a receptor-associated protein by binding-blocking and infection inhibition assay
To confirm the presence of receptor-associated proteins in the chromatography eluates isolated by S1N-SCZJ3, we attempted to block the binding of SCZJ3 to chicken embryo kidney cells (CEK) by pretreating virus with chromatography eluates, as shown in Fig 2A. Before infection, SCZJ3 virus was incubated with purified chromatography eluates of lung (L), kidney (K) and proventriculus (P). After infection, virus loads on CEK cells were determined by real-time PCR as described previously [1]. As shown in Fig. 2B, virus binding was efficiently blocked by lung and kidney chromatography elutes in a dose-dependent manner, which is consistent with the hypothesis that certain molecules in lung and kidney eluates comprise the virus receptor complex of IBV. However, the case of the proventriculus eluate is more complicated because certain molecules may enhance the binding of SCZJ3.
In order to determine whether the two proteins corresponding to the main bands (PDI and HSP70) in lung and kidney eluates are involved in the binding of SCZJ3, we examined whether these proteins could inhibit infection. Before infection, CEK cells were pretreated with anti-PDI and anti-HSP70 polyclonal antibody at different concentrations. As shown in Fig. 2C, reduction of virus load in CEK cells was observed after treatment with anti-HSP70 antibody. However, no significant reduction was detected in cells pretreated with anti-PDI antibody. These results indicate that HSP70 is essential for the binding of SCZJ3 to cells. The presence of HSP70 in chromatography eluates was confirmed by Western blot analysis (Fig. 2D)
Discussion
The spike (S) protein is a determinant of coronavirus tropism [13]. Retargeting the ectodomain of the protein can change the host tropism [22]. IBV isolates have been reported in recent years that not only affect the upper respiratory tract but also show broader tissue tropism and higher pathogenicity. The QX-like genotype is one of the epidemic IBV strains in China, according our epidemiology study based on analysis of the S1 gene. Distinctive pathological changes to the proventriculus by QX-like strains have been described [23]. Here, we demonstrate that the binding characteristics of recombinant S1N-SCZJ3 is accordant with virus tropism. In a recent study, a Y43H change in the ArkDPI S1 protein was shown to enhance binding to the trachea [24]. In comparison to S1N-M41, four main divergent regions were found in the S1N-SCZJ3 sequence (Fig 1A), and certain mutations may correlate with the binding properties of S1N-SCZJ3 and virus tropism.
A galactose-binding-like domain was found in the S1 protein between aa 137 and 230 by structure prediction (Phyre2). As shown in Fig. 1A, this domain is relatively conserved in S1N-SCZJ3 and S1N-M41 and may be important for the virus to maintain its ability to infect birds. However, the amino acid sequence from aa 19 to 36 is highly variable, and mutations may lead to conformational changes in S1N-SCZJ3 to S1N-M41. Two amino acid changes at the N-terminal spike protein of transmissible gastroenteritis virus result in the loss of enteric tropism [25]. In the case of SARS coronavirus, the structure of the RBD has been analyzed. The receptor-binding motif (RBM) that makes contacts with ACE2 has been identified at residues 424 to 494, and mutations in the RBM are critical for affinity or infectivity [26]. In IBV, a study of the conformation of the binding domains of different genotypes may help to identify the main binding sites that contact cellular receptors and investigate their correlation with tissue tropism.
Affinity chromatography is an efficient method to isolate virus receptors [19]. However, inhibition of binding and infection provides only indirect evidence for identifying virus receptors, and more-direct evidence is needed. However, BHK-21 cells transfected with one of the proteins described above were not sensitive to IBV infection (not shown), suggesting that the virus-receptor interaction of IBV is a multi-step process or that the infection is cell-type-specific. Certain molecules may be critical for virus reproduction in the cell [27]. Thus, reconstruction of the infection pathway of IBV will be more convenient if chicken-derived cell lines are used.
The presence of the S1N-SCZJ3 protein and other proteins in chromatography eluates complicates the analysis of the functions of these proteins. The functions of 14-3-3 and tropomyosin in virus infection need further study. These proteins may be important for infection of the proventriculus by SCZJ3. Surface-displayed HSP70 has been reported to be part of receptor complex in the case of dengue virus and Japanese encephalitis virus [17, 28]. HSP70 is a kind of stress protein that mainly induce ATP binding, and hydrolysis regulates its affinity for polypeptides. HSPs are important for folding of viral proteins [29]. HSP70 is probably upregulated by infection with a virus like avirulent NDV or by fever. This might help to explain the co-infections with IBV and other pathogens.
In conclusion, this study revealed that S1N-SCZJ3 is the receptor binding domain of SCZJ3 and that HSP70 is part of the receptor complex. Studying the interaction between S1N peptides and the receptor complex will allow us to better understand the infection process and pathogenesis of IBV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the State Natural Sciences Foundation (NSFC) (31372442, 31302094), the Project for Science and Technology Support Program of Sichuan Province (2014NZ0002, 2016NZ0003), the program of Main Livestock Standardized Breeding Technology Research and Demonstration (2016NYZ0052), and the Earmarked Fund for Modern Agroindustry Technology Research System (CARS-41-K09).
Compliance with ethical standards
Ethical statement
All procedures performed in studies involving animals were approved by the Animal Experimentation Ethics Committee of Sichuan University.
Conflict of interest
We declare that there is no conflict of interest.
Footnotes
Z. Zhang and X. Yang contributed equally to the research.
References
- 1.Fan WQ, Wang HN, Zhang Y, Guan ZB, Wang T, Xu CW, Zhang AY, Yang X. Comparative dynamic distribution of avian infectious bronchitis virus M41, H120, and SAIBK strains by quantitative real-time RT-PCR in SPF chickens. Biosci Biotechnol Biochem. 2012;76(12):2255–2260. doi: 10.1271/bbb.120521. [DOI] [PubMed] [Google Scholar]
- 2.Cook JK, Jackwood M, Jones RC. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- 3.Chu VC, McElroy LJ, Chu V, Bauman BE, Whittaker GR. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J Virol. 2006;80(7):3180–3188. doi: 10.1128/JVI.80.7.3180-3188.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 5.Peng G, Sun D, Rajashankar KR, Qian Z, Holmes KV, Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci. 2011;108(26):10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborti S, Prabakaran P, Xiao X, Dimitrov DS. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol J. 2005;2:73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Promkuntod N, van Eijndhoven REW, de Vrieze G, Grone A, Verheije MH. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Zhou Y, Wang H, Zeng F, Yang X, Zhang Y, Zhang A. Molecular detection and Smoothing spline clustering of the IBV strains detected in China during 2011–2012. Virus Res. 2016;211:145–150. doi: 10.1016/j.virusres.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Abro SH, Renstrom LHM, Ullman K, Belak S, Baule C. Characterization and analysis of the full-length genome of a strain of the European QX-like genotype of infectious bronchitis virus. Arch Virol. 2012;157(6):1211–1215. doi: 10.1007/s00705-012-1284-0. [DOI] [PubMed] [Google Scholar]
- 10.Yudong W, YongLin W, Zichun Z, GenChe F, Yihai J, Xiange L, Jiang D, Shushuang W. Isolation and identification of glandular stomach type IBV (QX IBV) in chickens. Chin J Anim Quar. 1998;15(1):1–3. [Google Scholar]
- 11.Benyeda Z, Szeredi L, Mato T, Suveges T, Balka G, Abonyi-Toth Z, Rusvai M, Palya V. Comparative histopathology and immunohistochemistry of QX-like, Massachusetts and 793/B serotypes of infectious bronchitis virus infection in chickens. J Comp Pathol. 2010;143(4):276–283. doi: 10.1016/j.jcpa.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abd El Rahman S, El-Kenawy AA, Neumann U, Herrler G, Winter C. Comparative analysis of the sialic acid binding activity and the tropism for the respiratory epithelium of four different strains of avian infectious bronchitis virus. Avian Pathol. 2009;38(1):41–45. doi: 10.1080/03079450802632049. [DOI] [PubMed] [Google Scholar]
- 13.Wickramasinghe IN, de Vries RP, Grone A, de Haan CA, Verheije MH. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol. 2011;85(17):8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madu IG, Chu VC, Lee H, Regan AD, Bauman BE, Whittaker GR. Heparan sulfate is a selective attachment factor for the avian coronavirus infectious bronchitis virus Beaudette. Avian Dis. 2007;51(1):45–51. doi: 10.1637/0005-2086(2007)051[0045:HSIASA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 15.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breslin JJ, Mork I, Smith MK, Vogel LK, Hemmila EM, Bonavia A, Talbot PJ, Sjostrom H, Noren O, Holmes KV. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37 C. J Virol. 2003;77(7):4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, Laxminarayana SV, Chandra N, Ravi V, Desai A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology. 2009;385(1):47–57. doi: 10.1016/j.virol.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol. 2005;79(8):4557–4567. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu P-w, Wu X, Wang H-n, Ma B-c, Ding M-d, Yang X (2015) Assembly and immunogenicity of baculovirus-derived infectious bronchitis virus–like particles carrying membrane, envelope and the recombinant spike proteins. Biotechnol Lett. 38(2):299–304. [DOI] [PMC free article] [PubMed]
- 21.M-d Ding, H-n Wang, H-p Cao, W-q Fan, Ma B-c Xu, P-w Zhang A-y, Yang X. Development of a multi-epitope antigen of S protein-based ELISA for antibodies detection against infectious bronchitis virus. Biosci Biotechnol Biochem. 2015;79(8):1287–1295. doi: 10.1080/09168451.2015.1025692. [DOI] [PubMed] [Google Scholar]
- 22.Kuo L, Godeke G-J, Raamsman MJ, Masters PS, Rottier PJ. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74(3):1393–1406. doi: 10.1128/JVI.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benyeda Z, Mato T, Suveges T, Szabo E, Kardi V, Abonyi-Toth Z, Rusvai M, Palya V. Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol. 2009;38(6):449–456. doi: 10.1080/03079450903349196. [DOI] [PubMed] [Google Scholar]
- 24.Leyson C, Franca M, Jackwood M, Jordan B. Polymorphisms in the S1 spike glycoprotein of Arkansas-type infectious bronchitis virus (IBV) show differential binding to host tissues and altered antigenicity. Virology. 2016;498:218–225. doi: 10.1016/j.virol.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballesteros M, Sanchez C, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227(2):378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 27.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4(5):e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vega-Almeida TO, Salas-Benito M, De Nova-Ocampo MA, del Angel RM, Salas-Benito JS. Surface proteins of C6/36 cells involved in dengue virus 4 binding and entry. Arch Virol. 2013;158(6):1189–1207. doi: 10.1007/s00705-012-1596-0. [DOI] [PubMed] [Google Scholar]
- 29.Hooper PL, Hightower LE, Hooper PL. Loss of stress response as a consequence of viral infection: implications for disease and therapy. Cell Stress Chaperones. 2012;17(6):647–655. doi: 10.1007/s12192-012-0352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


