Abstract
Recently, outbreaks associated with equine coronavirus (ECoV) have occurred in Japan and the United States. While ECoV is likely to be pathogenic to horses, it has not been shown that experimental inoculation of horses with ECoV produces clinical signs of disease. In this study, we inoculated three Japanese draft horses with an ECoV-positive diarrheic fecal sample to confirm infection after inoculation and to investigate the clinical course and virus shedding patterns of ECoV. Virus neutralization tests showed that all three horses became infected with ECoV. Two of the three horses developed clinical signs similar to those observed during ECoV outbreaks, including fever, anorexia, and gastrointestinal dysfunction. All horses excreted a large amount of virus into their feces for more than 9 days after inoculation regardless of the presence or absence of clinical signs, which suggests that feces are an important source of ECoV infection. ECoV was also detected in nasal swabs from all horses, suggesting that respiratory transmission of ECoV may occur. Both symptomatic horses developed viremia, while the asymptomatic horse did not. White blood cell counts and serum amyloid A concentrations changed relative to the clinical condition of the inoculated horses; these may be useful markers for monitoring the clinical status of horses infected with ECoV. This is the first report of induction of clinical signs of ECoV infection in horses by experimental inoculation. These clinical and virological findings should aid further investigation of the pathogenesis of ECoV.
Keywords: Fecal Sample, Nasal Swab, Clostridium Perfringens, Experimental Inoculation, Virus Neutralization Test
Introduction
Equine coronavirus (ECoV) belongs to the species Betacoronavirus 1 in the genus Betacoronavirus, family Coronaviridae; this species also includes bovine coronavirus (BCoV) [2]. ECoV (NC99 strain) was first isolated in the United States in 1999 from a foal with diarrhea [3]. There have been multiple outbreaks associated with ECoV at a racecourse in Japan [9, 12, 13], and in 2011–2012, outbreaks occurred in adult horses in the United States [14]. Recently, ECoV was detected in diarrheic and respiratory samples in Europe [8]. Fever, anorexia, lethargy, leukopenia and diarrhea were the main clinical signs observed in the horses involved in these outbreaks. ECoV is likely to be pathogenic to horses; however, experimental inoculation of horses with ECoV has not been reported to produce clinical signs of disease. An experimental model is necessary to understand the pathogenesis of ECoV. The objectives of this study were to confirm infection and to investigate the clinical course of disease and virus shedding patterns in horses experimentally inoculated with ECoV.
Materials and methods
Horses
Three 9- to 10-month-old Japanese draft horses were used in this study. All were healthy and had no serological evidence of prior ECoV infection (based on virus neutralization tests; antibody titers <8 for antibodies to the NC99 and Obihiro12-2 strains) (Table 1). The NC99 and Obihiro12-2 strains were isolated from a foal and an adult horse that presented with diarrhea in the United States [3] and Japan [13], respectively.
Table 1.
Virus neutralization titers to two equine coronavirus strains, NC99 and Obihiro12-2
| NC99 | Obihiro12-2 | |||
|---|---|---|---|---|
| 0 dpi | 14 dpi | 0 dpi | 14 dpi | |
| Horse 1 | <8 | 128 | <8 | 64 |
| Horse 2 | <8 | 128 | <8 | 128 |
| Horse 3 | <8 | 32 | <8 | 32 |
dpi, days post-inoculation
Inoculum
An ECoV-positive sample of watery diarrheic feces collected from a 4-year-old female draft horse during an ECoV outbreak in 2012 [13] was used as the inoculum. The horse had watery diarrhea for 5 days prior to collection of the sample. Virus neutralization tests on paired serum samples collected from this horse showed seroconversion against ECoV. The ECoV-positive sample was stored at −80 °C until inoculum preparation. We attempted to isolate ECoV from the sample using HRT-18G cells as described previously [12], but were unsuccessful. Prior to administration, the sample was tested for other pathogens associated with intestinal disease, including equine rotavirus, Clostridium perfringens, Clostridium difficile and Salmonella species. Testing for equine rotavirus was performed using reverse transcription loop-mediated isothermal amplification [10]. Previously described culture methods were used to screen for Clostridium perfringens, Clostridium difficile and Salmonella species [11]. The inoculum was negative for equine rotavirus, Clostridium difficile and Salmonella species, but Clostridium perfringens was isolated from the sample. The sample was diluted 1:10 in phosphate-buffered saline (PBS), and 1000 ml of this 10 % fecal suspension was administered into the esophagus of each experimental horse using a transnasal catheter while under sedation. The suspension contained 4.2 × 109 copies/head of the ECoV nucleocapsid gene; the method used to determine copy numbers is described later in this section.
Sample collection
Clinical examinations were performed daily, and rectal temperatures were measured twice daily during the 14 days post-inoculation (dpi). Horses with rectal temperatures exceeding 38.6 °C were defined as significantly pyretic. Feces, nasal swabs, serum and EDTA blood samples were collected from horses at 0–8, 10, 12 and 14 dpi. Fecal samples were diluted 1:10 in Dulbecco’s modified Eagle’s medium supplemented with 100 units of penicillin, 100 μg of streptomycin, 50 μg of gentamicin and 0.25 μg of amphotericin B per ml. Fecal suspensions were clarified by centrifugation at 2000×g for 10 min prior to use in real-time reverse transcription polymerase chain reaction (real-time RT-PCR). Horses 1 and 2 presented with clinical signs at 2–8 and 2–6 dpi, respectively; feces collected from these horses during their period of clinical disease were diluted 1:10 in PBS and used for bacterial isolation. Nasal swabs were collected using 1.0 cm × 1.5 cm absorbent cotton swabs and were immersed in 2.5 ml of PBS supplemented with 0.6 % tryptose phosphate broth and 500 units of penicillin, 500 μg of streptomycin and 1.25 μg of amphotericin B per ml. White blood cell counts were performed on the EDTA blood samples using an automatic analyzer (MEK-6450 Celltac alpha, Nihon Kohden Corp., Tokyo, Japan). Leukopenia and lymphopenia were defined as white blood cell and lymphocyte counts of less than 4500 and 1600 cells/μl, respectively. The experimental protocol and all animal procedures were approved by the Animal Care Committee of the Equine Research Institute of the Japan Racing Association.
Real-time RT-PCR
Viral RNA was extracted from 100-μl samples of the fecal suspensions (equivalent to 10 mg of feces), nasal swabs, and EDTA blood samples (equivalent to 100 μl of nasal swab or blood sample) using a nucleic acid isolation kit (MagNA Pure LC Total Nucleic Acid Isolation Kit, Roche Diagnostics GmbH, Mannheim, Germany). Viral RNA was eluted using elution buffer (100 μl) and stored at −80 °C prior to use. Real-time RT-PCR was performed using a previously described primer set [14] and TaqMan Fast Virus 1-Step Master Mix (Life Technologies, Carlsbad, CA, U.S.A.) according to the manufacturer’s instructions. Real-time RT-PCR was performed in 20-µl reaction mixtures containing 4 μl of template, which was control RNA or extracted fecal samples (equivalent to 0.4 mg of feces) or nasal swabs or blood samples (equivalent to 4 μl of nasal swab or blood sample). Thermal cycling conditions included an initial hold at 50 °C for 5 min, 95 °C for 20 s, and then 40 cycles at 95 °C for 3 s and 60 °C for 30 s.
To create a standard curve for the real-time RT-PCR reaction, control ECoV RNA was synthesized as described previously [10]. In brief, a DNA fragment including the nucleocapsid gene was generated from viral RNA prepared from the NC99 strain by conventional RT-PCR using the primer set for ECoV-Nf and ECoV-Nr [12]. The product obtained in the first amplification was used as a template for the second amplification, which was performed using the primer pair T3-ECoV-Nf (5′-attaaccctcactaaagggagaatgtctttcactcctggcaagc-3′), containing the T3 promoter sequence, and ECoV-Nr. RNA was synthesized using T3 RNA polymerase, and the RNA copy number was calculated from its absorbance value at 260 nm. Real-time RT-PCR was performed twice on triplicate samples of tenfold serial dilutions of control RNA (2 × 108-2 × 100 copies/4 μl). The results indicated that the real-time RT-PCR could reliably detect as few as 200 copies/reaction. In contrast, at 20 copies/reaction, only five out of six samples were positive for ECoV, and the average Ct value of the five positive samples was 36.0 cycles. Detection after 35.0 cycles is less reliable, and therefore, we decided for the purpose of this study that real-time RT-PCR could reliably detect a minimum of 200 copies/reaction. Two hundred copies per reaction corresponds to 5.0 × 105 copies per gram in feces and 5.0 × 104 copies per milliliter in nasal swabs and blood samples. All real-time RT-PCR reactions were performed in duplicate and average copy numbers were calculated.
Virus neutralization test
The virus neutralization test for ECoV was performed on paired serum samples collected at 0 and 14 dpi, and on HRT-18G cells as described previously [12]. The NC99 and Obihiro12-2 strains were used as the challenge viruses. Twofold serial dilutions of serum were mixed with an equal volume of viral suspensions containing two hundred 50 % tissue culture infective doses per 0.1 ml and incubated for 60 min at 37 °C. The mixtures were applied to HRT-18G cells and incubated for 5–7 days. Virus-neutralizing antibody titers were expressed as the reciprocal of the highest serum dilution that inhibited viral cytopathic effects. Seroconversion was defined as a 4-fold or greater increase in antibody titers between paired serum samples.
Measurement of serum amyloid A
Serum amyloid A (SAA) concentrations were measured to monitor the inflammatory response to viral inoculation. SAA was measured by a latex agglutination method using the LZ test ‘EIKEN’ SAA (Eiken Chemical Co., Ltd., Shimotsuga-gun, Japan) and an automated analyzer (Hitachi 7020, Hitachi High-Technologies Corporation, Tokyo, Japan) [4]. SAA concentrations of more than 20 µg/ml were defined as abnormally high, as described by Belgrave et al. [1].
Results
Horses 1 and 2 developed biphasic fevers from 2 to 4 dpi, whereas horse 3 did not become febrile during the observation period; the rectal temperatures of the horses are shown in Fig. 1. Horses 1 and 2 were also anorexic at 2–4 and 4–6 dpi, and excreted pasty feces at 2–8 and 6 dpi, respectively. In contrast, horse 3 did not develop any clinical signs during the study period. Horse 1 became leukopenic (Fig. 2) and lymphopenic at 2–4 dpi, and at 2 and 4 dpi, respectively; however, although the white blood cell counts of horses 2 and 3 tended to decrease, they did not reach levels sufficiently low to be considered leukopenic. The SAA concentrations of horses 1 and 2 were abnormally high for 7 and 4 days, and peaked at 3 and 4 dpi, respectively (Fig. 3). The fecal samples collected at 2–8 (horse 1) and 2–6 (horse 2) dpi were negative for Clostridium difficile, Clostridium perfringens, and Salmonella species.
Fig. 1.
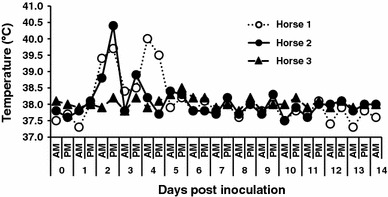
Rectal temperatures of inoculated horses
Fig. 2.
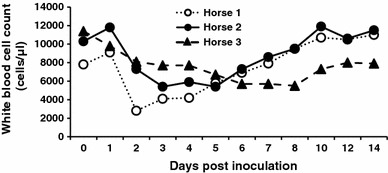
White blood cell counts of inoculated horses
Fig. 3.
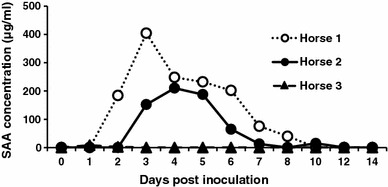
Serum amyloid A (SAA) concentrations in inoculated horses
All horses began to shed ECoV RNA in their feces at 3 or 4 dpi and continued shedding the virus until 12 or 14 dpi. The amount of ECoV RNA in the fecal samples peaked at 7 or 8 dpi, and peak copy numbers ranged from 2.7 × 109 to 5.5 × 1010 copies/g (Fig. 4A). ECoV RNA was detected in nasal swabs from horses 1, 2 and 3 at 6–10, 5–14 and 7–10 dpi, respectively. The ECoV RNA copy numbers obtained from nasal swabs from horses 1, 2 and 3 reached maximum values of 2.2 × 105, 1.0 × 106 and 1.4 × 105 copies/ml at 6, 6 and 7 dpi, respectively (Fig. 4B). ECoV RNA was detected in EDTA blood samples from horses 1 and 2 from 2–7 and 2 dpi, respectively. At 2 dpi, the ECoV RNA in EDTA blood samples reached maximum copy numbers of 1.3 × 106 (horse 1) and 1.2 × 105 (horse 2) copies/ml. ECoV RNA was not detected in EDTA blood from horse 3 at any time during this study (Fig. 4C).
Fig. 4.
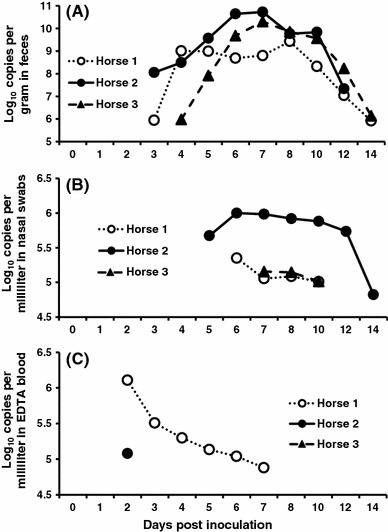
Detection of ECoV in fecal samples (A), nasal swabs (B), and EDTA blood samples (C) of inoculated horses by real-time RT-PCR. The log-transformed (base 10) values of ECoV gene copies per gram or milliliter are shown. Samples that had less than 5.0 × 105 (105.7) copies/g in feces and 5.0 × 104 (104.7) copies/ml in nasal swabs and EDTA blood samples were considered negative for ECoV
Virus neutralization titers revealed that all horses seroconverted to both NC99 and Obihiro12-2 (Table 1).
Discussion
To our knowledge, this is the first report of successful reproduction of the clinical signs of ECoV after experimental inoculation in horses. In Japan, ECoV outbreaks occurred among Japanese draft horses at the same racecourse in 2004, 2009 and 2012 [9, 12, 13]. We previously attempted to reproduce the clinical signs of ECoV infection by experimental administration of cultured ECoVs to Thoroughbred horses but failed to induce clinical symptoms of disease [Nemoto, unpublished data]. However, BCoV is closely related to ECoV, and experimental inoculation with BCoV-positive fecal samples or intestinal contents has reproduced the clinical signs of BCoV infection in several experimental challenge studies [15–17]. In addition, ECoV outbreaks in Japan have been reported only in Japanese draft horses. Therefore, to reproduce the clinical signs caused by ECoV infection experimentally, we inoculated Japanese draft horses with an ECoV-positive fecal sample.
Two of the three horses in this challenge study developed clinical signs similar to those seen during ECoV outbreaks, including anorexia, pyrexia and soft feces. Although Clostridium perfringens was isolated from the inoculum, it was not isolated from the fecal samples collected during the days on which the two horses showed clinical signs. This suggests that Clostridium perfringens was not related to the clinical symptoms of disease. The results of serological testing indicated that all horses were infected with ECoV; however, one horse showed no signs of illness, suggesting that subclinical ECoV infection can occur in some horses. None of the horses in this study developed severe watery diarrhea similar to that of the horse from which the inoculum was obtained. This is consistent with previous reports that diarrhea was observed in only some of the affected horses during ECoV outbreaks [12–14], and these findings suggest that additional factors may be associated with induction of diarrhea in ECoV-infected horses.
Isolation of ECoV in cell culture is difficult [7], and therefore, real-time RT-PCR was used to investigate virus shedding patterns in the horses infected with ECoV in this study. The two symptomatic horses became viremic, while the asymptomatic horse did not. Leukopenia and lymphopenia were observed only in horse 1, which had a viral load of more than 106 copies/ml in EDTA blood; the white blood cell count of this horse increased as the ECoV load decreased. White blood cell count has been shown to be closely related to clinical symptoms in horses infected with ECoV. SAA, which is used as a marker of inflammation in equine clinical practice because it closely reflects changes in clinical conditions [5], increased in the two symptomatic horses in concordance with clinical signs. Thus, white blood cell count and SAA may both be useful markers of ECoV infection in horses.
Real-time RT-PCR of fecal samples detected a large amount of virus in the feces for more than 9 days in all horses regardless of presence or absence of clinical signs. These results indicate that ECoV replicates in the intestinal tract and that feces are an important source of ECoV infection. Irrespective of their symptomatic or asymptomatic status, infected horses are capable of spreading the virus into the environment. Because of the large amount of virus shed into the feces, the main transmission route of ECoV is likely fecal-oral.
ECoV was also detected in nasal swabs from all horses, even though the inoculum was administered directly into the esophagus. In addition, the virus was shed from the nasal tracts of the two horses that developed fever even after the horses became afebrile. In Europe, ECoV was also detected from a respiratory specimen within a few days after the onset of clinical signs [8]. In previous studies using field samples from the outbreaks in 2009 and 2012 [12, 13], ECoV was not detected in nasal swabs. This may be because the nasal swabs were collected during the early stages of fever development in these previous reports. As ECoV was not inoculated via the respiratory route in our current study, viral infection of the respiratory tract was probably secondary to the viremia that resulted after infection of the intestinal tract. However, in horse 3, viremia was not detected, but ECoV was isolated from nasal swabs. Therefore, we hypothesize that another route, such as lymphogenous transmission, may be related to the spread of the ECoV within the host. BCoV, which is closely related to ECoV, is transmitted to other cattle via both the respiratory and the fecal-oral route [6], and it is therefore possible that respiratory transmission of ECoV may also occur. Alternatively, it is possible that the nasal tract was secondarily infected with ECoV via exposure to feces on the floor of the stall. To clarify the route of transmission of ECoV, future virus challenge studies using respiratory tract inoculation should be performed.
We successfully reproduced the clinical signs of ECoV infection by experimental inoculation of an ECoV-positive fecal sample into the gastrointestinal tracts of draft horses. The clinical and virological findings reported here should aid further studies of the pathogenesis of ECoV.
Acknowledgments
The equine coronavirus NC99 strain was kindly provided by Dr. J. S. Guy of North Carolina State University, Raleigh, NC, USA. We are grateful to Ms. Ryoko Ishihara (Hokkaido Research Station, National Institute of Animal Health, Sapporo, Japan) and Dr. Chihiro Yoshihara (RIKEN Brain Science Institute, Wako, Japan) for their invaluable suggestions, and to Mr. Akira Kokubun, Ms. Kaoru Makabe, Ms. Akiko Suganuma, and Ms. Kazue Arakawa (Epizootic Research Center, Equine Research Institute, Japan Racing Association) for their invaluable technical assistance.
References
- 1.Belgrave RL, Dickey MM, Arheart KL, Cray C. Assessment of serum amyloid A testing of horses and its clinical application in a specialized equine practice. J Am Vet Med Assoc. 2013;243:113–119. doi: 10.2460/javma.243.1.113. [DOI] [PubMed] [Google Scholar]
- 2.de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebunhr J. Coronaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. London: Elsevier; 2011. pp. 806–828. [Google Scholar]
- 3.Guy JS, Breslin JJ, Breuhaus B, Vivrette S, Smith LG. Characterization of a coronavirus isolated from a diarrheic foal. J Clin Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobo S, Niwa H, Anzai T. Evaluation of serum amyloid A and surfactant protein D in sera for identification of the clinical condition of horses with bacterial pneumonia. J Vet Med Sci. 2007;69:827–830. doi: 10.1292/jvms.69.827. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsen S, Andersen PH. The acute phase protein serum amyloid A (SAA) as a marker of inflammation in horses. Equine Vet Educ. 2007;19:38–46. doi: 10.1111/j.2042-3292.2007.tb00550.x. [DOI] [Google Scholar]
- 6.Kanno T. Bovine coronavirus infection: pathology and interspecies transmission. J Disaster Res. 2012;7:297–302. [Google Scholar]
- 7.Magdesian KG, Dwyer RM, Arguedas MG. Viral Diarrhea. In: Sellon DC, Long MT, editors. Equine infectious diseases. 2. St. Louis: Elsevier; 2014. pp. 198–203. [Google Scholar]
- 8.Miszczak F, Tesson V, Kin N, Dina J, Balasuriya UB, Pronost S, Vabret A. First detection of equine coronavirus (ECoV) in Europe. Vet Microbiol. 2014;171:206–209. doi: 10.1016/j.vetmic.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narita M, Nobumoto K, Takeda H, Moriyama T, Morita Y, Nakaoka Y. Prevalence of disease with inference of equine coronavirus infection among horses stabled in a draft-horse racecourse. J Jpn Vet Med Assoc. 2011;64:535–539. [Google Scholar]
- 10.Nemoto M, Imagawa H, Tsujimura K, Yamanaka T, Kondo T, Matsumura T. Detection of equine rotavirus by reverse transcription loop-mediated isothermal amplification (RT-LAMP) J Vet Med Sci. 2010;72:823–826. doi: 10.1292/jvms.09-0446. [DOI] [PubMed] [Google Scholar]
- 11.Niwa H, Kato H, Hobo S, Kinoshita Y, Ueno T, Katayama Y, Hariu K, Oku K, Senoh M, Kuroda T, Nakai K. Postoperative Clostridium difficile infection with PCR ribotype 078 strain identified at necropsy in five Thoroughbred racehorses. Vet Rec. 2013;173:607. doi: 10.1136/vr.101960. [DOI] [PubMed] [Google Scholar]
- 12.Oue Y, Ishihara R, Edamatsu H, Morita Y, Yoshida M, Yoshima M, Hatama S, Murakami K, Kanno T. Isolation of an equine coronavirus from adult horses with pyrogenic and enteric disease and its antigenic and genomic characterization in comparison with the NC99 strain. Vet Microbiol. 2011;150:41–48. doi: 10.1016/j.vetmic.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oue Y, Morita Y, Kondo T, Nemoto M. Epidemic of equine coronavirus at Obihiro racecourse, Hokkaido, Japan in 2012. J Vet Med Sci. 2013;75:1261–1265. doi: 10.1292/jvms.13-0056. [DOI] [PubMed] [Google Scholar]
- 14.Pusterla N, Mapes S, Wademan C, White A, Ball R, Sapp K, Burns P, Ormond C, Butterworth K, Bartol J, Magdesian G. Emerging outbreaks associated with equine coronavirus in adult horses. Vet Microbiol. 2013;162:228–231. doi: 10.1016/j.vetmic.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saif LJ, Redman DR, Moorhead PD, Theil KW. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am J Vet Res. 1986;47:1426–1432. [PubMed] [Google Scholar]
- 16.Traven M, Naslund K, Linde N, Linde B, Silvan A, Fossum C, Hedlund KO, Larsson B. Experimental reproduction of winter dysentery in lactating cows using BCV—comparison with BCV infection in milk-fed calves. Vet Microbiol. 2001;81:127–151. doi: 10.1016/S0378-1135(01)00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsunemitsu H, Smith DR, Saif LJ. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch Virol. 1999;144:167–175. doi: 10.1007/s007050050493. [DOI] [PMC free article] [PubMed] [Google Scholar]


