Abstract
We previously determined the crystal structure of the wild-type keratin 1/10 helix 2B heterodimer at 3.3 Å resolution. We proposed that the resolution of the diffraction data was limited due to the crystal packing effect from keratin 10 (K10) residue Cys401. Cys401K10 formed a disulfide-linkage with Cys401 from another K1/10 heterodimer, creating an “X-shaped” structure and a loose crystal packing arrangement. We hypothesized that mutation of Cys401K10 to alanine would eliminate the disulfide-linkage and improve crystal packing thereby increasing resolution of diffraction and enabling a more accurate side chain electron density map. Indeed, when a K10 Cys401Ala 2B mutant was paired with its native keratin 1 (K1) 2B heterodimer partner its x-ray crystal structure was determined at 2.07 Å resolution; the structure does not contain a disulfide linkage. Superposition of the K1/K10(Cys401Ala) 2B structure onto the wild-type K1/10 2B heterodimer structure had a root-mean-square-deviation of 1.88 Å; the variability in the atomic positions reflects the dynamic motion expected in this filamentous coiled-coil complex. The electrostatic, hydrophobic, and contour features of the molecular surface are similar to the lower resolution wild-type structure. We postulated that elimination of the disulfide linkage in the K1/K10(Cys401Ala) 2B structure could allow for the 2B heterodimers to bind/pack in the A22 tetramer configuration associated with mature keratin intermediate filament assembly. Analysis of the crystal packing revealed a half-staggered anti-parallel tetrameric complex of 2B heterodimers; however, their register is not consistent with models of the A22 mode of tetrameric alignment or prior biochemical cross-linking studies.
Keywords: keratin, structure, intermediate filament, heterodimer, disulfide bond, assembly
Introduction
Intermediate filaments (IFs) are a fundamental fibrous component of the cytoskeleton and have an essential role in human health and disease: mutations in IF proteins cause or predispose humans to more than 80 tissue-specific diseases, collectively called “IF-pathies” [1]. IFs are one of three major filamentous structures comprising the cytoskeleton–actin microfilaments, IFs, and microtubules. IFs are named such because their diameter of 10-nm is between the diameter of microfilaments (~6-nm) and microtubules (~25-nm) [2]. IFs regulate many cellular activities: physical support, intracellular signaling, protein scaffolding, protein transport, and cellular stress [3-5]. IFs also are increasingly linked to an active role in cancer pathogenesis, including cancer cell invasion and metastasis [6,7].
IF proteins are divided into six types, the most abundant being types I and II−they represent 54 different keratins which can be epidermal, trichocyte, or epithelial [1,3,4]. Keratins form heteropolymers, with one type I and one type II keratin partnering to form a heterodimer early in the filament assembly process. Type III IFs, including vimentin, desmin, and glial fibrillary acidic protein (GFAP), differ from keratins because they form homopolymers. Type IV IFs are the neurofilament proteins; type V IFs are the nuclear lamins; and type VI IFs are eye lens proteins (Figure 1).
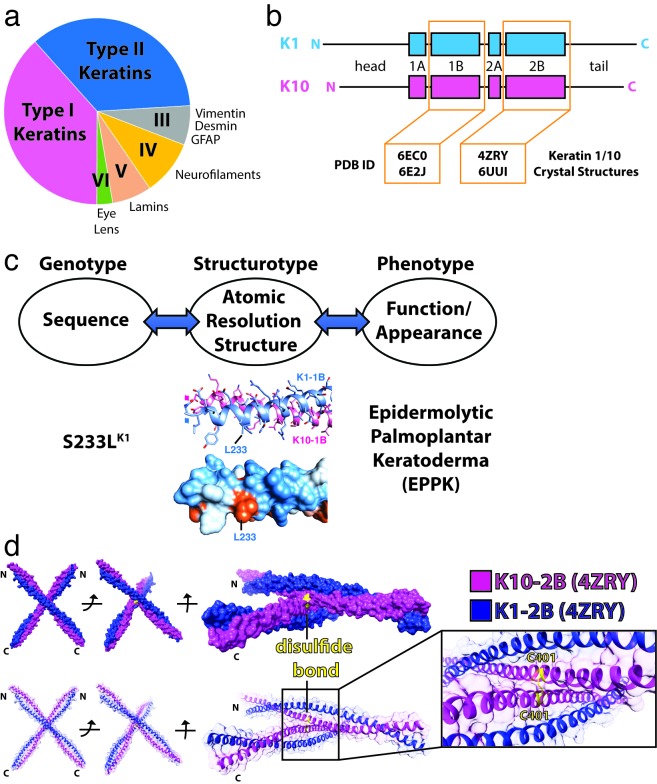
Figure 1.
Intermediate filaments and the structural biology of keratin 1/10 complex. (a) The six major types of intermediate filaments, with the area of each pie slice representative of the total number of IFs for that type. (b) Common structural organization of IFs using K1 and K10 as an example, with the head, tail, and four helical domains denoted. Protein Data Bank (PDB) ID codes for determined K1/K10 crystal structures are accentuated (orange). (c) Proposed paradigm for correlating genotype, structurotype, and phenotype for clinical diseases. An example using the K1S233L mutation causing epidermolytic palmoplantar keratoderma (EPPK) is shown. L233 creates aberrant hydrophobicity (orange) on the molecular surface of K1S233L/K10-1B (PDB ID 6E2J). (d) Crystal structure of the wild-type K1/K10-2B heterodimer (PDB ID 4ZRY) depicting that a K10C401-mediated disulfide bond (yellow) occurs between K10 helices on separate heterodimers.
All IF proteins share a common structural organization: a central coiled-coil (historically divided into four helical domains: 1A, 1B, 2A, 2B) flanked on either end by head and tail domains of varying lengths and composition (Figure 1b). The keratin heads and tails, for example, contain glycine-rich sequences predicted to form “glycine loops” [8]. They play an important role in the tissue-specific functions of keratins, in part because the head and tail domains are the primary location for phosphorylation and other post-translational modifications that regulate IF structure and function [9]. The atomic resolution structure of keratin head and tail domains remains unknown; however, one molecular modeling study hypothesized that “aromatic zippers” interact to form a tetrameric terminal domain complex [10]. While domains 2A and 2B (and their interspacing linker, L2) have been historically separated, they are now often referred to as “coil 2” based on the identification of hendecad repeats in 2A and L2 and a fully-helical vimentin coil 2 crystal structure [11,12]. Here, we use the historical nomenclature because it facilitates our ability to correlate previous biochemical studies on modes of IF assembly (A11, A22, A12, ACN) with newly identified structural mechanisms [13].
To date there are six crystal structures of epidermal keratin heterocomplexes deposited in the Protein Data Bank (PDB): four K1/K10 and two K5/14 (Table 1). Each structure has added new molecular insights into the function of keratin IFs, thus emphasizing the critical value that each experimentally-determined structure has for elucidating the mechanisms of action of IF proteins. All of the structures are of either the 1B or 2B helical domains of the central coiled-coil; there are no keratin structures capturing the 1A or 2A helical domains. Important questions in keratin biology, as well as for IF biology as a whole, are why so many different IF proteins exist and how do they generate their functional diversity despite sharing a common coiled-coil structural fold? Our wild-type K1/K10-2B structure (PDB ID 4ZRY) provided some insight into these questions: it demonstrated that only a few amino acid sequence differences are needed to significantly change the chemical properties of the IF molecular surface because, at least for K1/K10-2B, the unique residues were concentrated along the outer helical ridge of the coiled-coil [14]. The concept of “maximally diverse surface chemistry” offers one way that IFs may achieve a range of tissue-specific functions; it also emphasizes why investigation of the structural biology of each IF system is needed in order to properly understand IF functional differences.
Table 1. X-ray crystal structures determined of human keratin complexes.
| PDB ID & Reference | Resolution | Keratin Complex | Mutation | Domain (Residues) | Physiologic Assembly State | Key Structural Findings |
| 6EC0 [19] | 2.98 Å | K1-K10 | None | 1B (226-331 / 195-296) | A11 heterotetramer | Symmetric knob-pocket interactions in 1B domain are important for proper tetramer and mature IF assembly |
| 6E2J [19] | 2.39 Å | K1-K10 | K1-S233L | 1B (226-331 / 195-296) | A11 heterotetramer | Pathologic K1 mutation S233L creates aberrant hydrophobic interactions leading to keratin aggregation |
| 4ZRY [14] | 3.30 Å | K1-K10 | None | 2B (386-489 / 347-456) | Heterodimer (dimer was disulfide-linked into X-shaped tetramers) | Transdimer homotypic disulfide bond linkage at K10 position C401; a few residue differences significantly alter shape and chemistry of keratin molecular surface because unique residues often align along outer helical ridges of coiled-coil |
| 6UUI This report | 2.07 Å | K1-K10 | K10-C401A | 2B (383-489 / 349-456) | Heterodimer | Highest resolution keratin structure to date; an acidic 2B C-terminus drives an octameric complex of half-staggered anti-parallel 2B heterodimers and a splayed C-terminal tetrameric interface |
| 3TNU [23] | 3.01 Å | K5-K14 | None | 2B (382-476 / 332-421) | Heterodimer (dimer was disulfide-linked into X-shaped tetramers) | Transdimer homotypic disulfide bond linkage at K14 position C367; inter-IF disulfide linkages enable filament cage around nucleus to regulate nuclear shape |
| 6JFV [36] | 2.60 Å | K5-K14 | K14-C367A | 2B (379-476 / 327-421) | Heterodimer | Crystal lattice contacts reflect helix 2B-2B interactions important to proper IF assembly, possibly at the octamer stag |
Perhaps the most critical unanswered question in IF biology is how IF proteins assemble into mature 10-nm filaments−specifically, the molecular and structural interactions that dictate how they assemble. Using keratin IFs (KIFs) as an example, a wealth of biophysical studies have defined the stages of KIF assembly as: one type I keratin and one type II keratin pair to form a parallel heterodimer; heterodimers then bind to form an anti-parallel tetramer; eight tetramers then merge to form a protofibril/unit-length filament; and finally four protofibrils assemble into the complete KIF [15-17]. Similar biophysical studies have defined IF assembly for vimentin and other IF proteins and together these biophysical studies from multiple laboratories represent significant advances in understanding of IF function. However, it has been stated that “The most pressing current obstacle in the IF field is that there isn’t a single direct cure or even partial therapy for any of the human IF-pathies” [18]. We believe this predicament exists largely because of a single major knowledge gap–the absence of atomic resolution structures that explain in mechanistic detail how IF proteins assemble into filaments. This lack of knowledge hinders our explanation of how pathologic mutations alter IF protein structure and filament formation.
Toward the goal of determining IF assembly mechanisms in atomic resolution detail, we recently reported two K1/K10-1B crystal structures that captured the physiologic A11 tetrameric state of K1/K10-1B (PDB ID 6EC0 and 6E2J) [19]. K1 and K10 are the primary epidermal keratins expressed in suprabasal (spinous layer) keratinocytes. These structures led to the discovery of a knob-pocket tetramer assembly mechanism that was validated using light scattering and electron microscopy (EM) analyses of K1, K8, and vimentin knob mutants. The importance of this knob-pocket interaction to the stability of the IF tetramer, the building block of mature 10-nm filaments, is reflected by its structural conservation across type II keratins, type III IFs (e.g. vimentin, GFAP), and type V IFs (e.g. lamin A/C) (Figure 2).
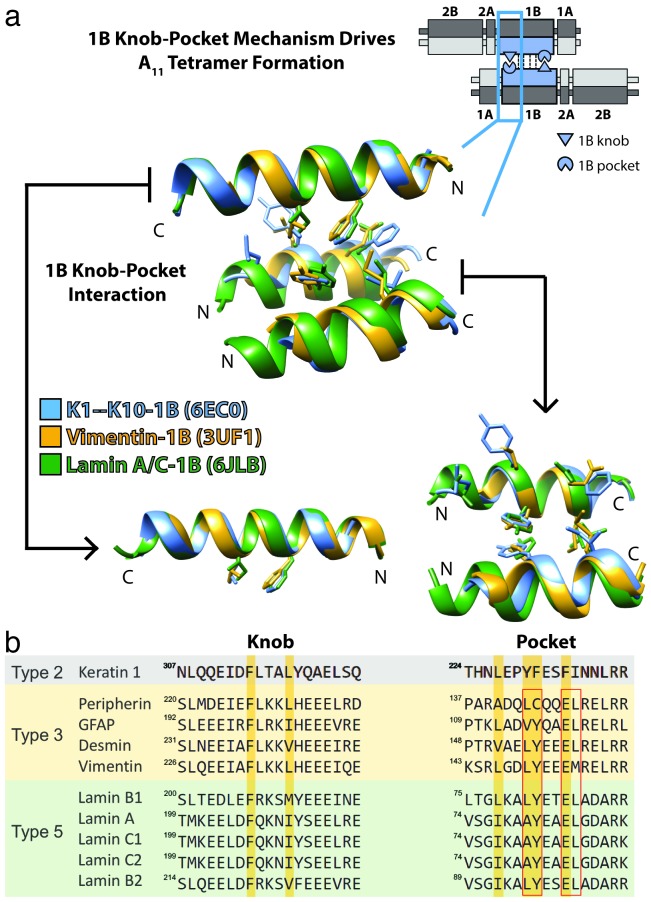
Figure 2.
Knob-pocket mechanism in helix 1B is important for A11 tetramer formation for multiple IF types. (a) For orientation, a schematic depicts a keratin A11 tetramer (upper right) and shows the position of symmetric knob-pocket interactions in the 1B helical region of the Type II keratin (blue). Below, the crystal structures of K1/K10-1B (blue, PDB ID 6EC0), vimentin-1B (gold, PDB ID 3UF1), and lamin A/C-1B (green, PDB ID 6JLB) were superposed and the knob-pocket interaction presented. There is structural conservation of the 1B knob-pocket tetramer assembly mechanism for Type II, Type III, and Type V IFs. There are no structures to date of Type IV or VI IFs. (b) Multiple sequence alignment for the knob and pocket regions of select Type II, Type III, and Type V IFs. There is complete conservation of the key phenylalanine residue in the knob. There is also a high degree of conservation of the pocket residues between Type III and Type V IFs (red boxes), which can be compared to the yellow bars for keratin 1. The structural superposition in panel (a) demonstrates the pocket is structurally conserved despite there being a variation in how the pocket is formed between heterodimeric IFs (Type I/II) and homodimeric IFs (Types III and V). The fact that heterodimeric and homodimeric IFs both utilize the 1B knob-pocket mechanism reinforces its importance to IF assembly.
In addition, one of these two K1/K10-1B structures contained a S233L mutation on K1 (PDB ID 6E2J), representing the only keratin structure to date to incorporate a mutation pathologic to humans. K1S233L causes epidermolytic palmoplantar keratoderma [20,21]. Determining structures of IF proteins that incorporate clinically-relevant human mutations is necessary to understand how errors in IF biology cause human disease. Traditionally, science and medicine has discussed “genotype-phenotype” correlation for human diseases. We believe that atomic resolution structure is necessary to fully correlate genotype with phenotype for any given disease; therefore, the paradigm that we propose is a “genotype-structurotype-phenotype” correlation for all mutations causing human pathology (Figure 1c). This level of knowledge about a disease can strengthen efforts to develop precision therapeutics by enabling the specific targeting of a structural mechanism that is responsible for a clinical phenotype.
One mutational “hotspot” in keratins is the C-terminus of helix 2B, where a conserved TYR*LLEGE sequence motif important to proper 10-nm filament assembly exists [22]. Crystal structures of K5/K14-2B (K5 and K14 are the primary keratins expressed in basal layer keratinocytes) and K1/K10-2B have been determined (PDB ID 3TNU and 4ZRY, respectively). They enabled the molecular modeling of mutations associated with several human diseases: epidermolysis bullosa simplex, epidermolytic ichthyosis, cyclic ichthyosis with epidermolytic hyperkeratosis, and epidermolytic and non-epidermolytic palmoplantar keratosis [23,24]. These modeling analyses demonstrated that some mutations alter the heterodimer interface, whereas others are surface-exposed and therefore are likely to alter higher-order filament assembly. Molecular details about how pathologic mutations affect 10-nm filament assembly remain elusive both by crystallography and cryo-EM.
An unexpected finding from the 2B crystal structures was an X-shaped superstructure. Both the K5/K14-2B and K1/K10-2B structures displayed a transdimer homotypic disulfide bond between type I keratins (K10, K14) (Figure 1d) [23,24]. For K5/K14, this disulfide bonding was shown to be physiologically relevant, since it regulated the dynamics and organization of keratin filament networks within the cell, especially in the perinuclear region [23,25]. It is important to recognize this disulfide linkage in vivo occurs between mature 10-nm filaments, not within an individual filament. One consequence of the disulfide linkage and X-shaped structure was a large solvent content in the K1/K10-2B crystal (~79%), which we believed limited the diffraction resolution to ~3.3-3.5 Å. We hypothesized that mutation of Cys401K10 to alanine would eliminate the disulfide bond and lead to higher diffraction resolution by improving crystal packing and reducing solvent content. Furthermore, we hypothesized that the 2B domains in the absence of the disulfide linkage would now be able to pack together in their purported physiologic A22 tetrameric state [13,26]. The four proposed modes of keratin heterodimer alignment into tetramers (A11, A22, A12, ACN) were discussed and illustrated in our A11 K1/K10-1B tetrameric structure manuscript [19].
Building from our wild-type K1/K10-2B structure, we report here the x-ray crystal structure of the complex between the helix 2B domains of human K1 and K10C401A. The structure demonstrated that the C401A substitution in K10 did in fact eliminate the transdimer homotypic disulfide bond observed in the wild-type structure. Multi-angle light scattering (MALS) confirmed K1/K10C401A-2B is a heterodimer in solution. Analysis of the crystal lattice packing identified an octameric assembly of K1/K10C401A heterodimers arranged in “half-staggered” and “splayed” configurations. The contact interfaces within the octameric assembly are largely driven by molecular interactions at the C-terminus of helix 2B, a hotspot for clinically relevant human mutations. The tetramer alignments within the crystal lattice did not correlate with the previously described A22 mode of keratin tetramer alignment. Together, this work addresses knowledge gaps in the structural biology of human KIFs and the molecular properties of helix 2B that are important for 10-nm filament assembly.
Materials and Methods
Plasmids
cDNA for wild-type human K1-2B (residues 370-489) and human K10-2B (residues 337-456) was generated as previously described [14]. K10C401A-2B was prepared using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) and the following primers: 5’- GCAGAAACAGAAGGTCGCTACGCGGTGCAGCTCTCACAGATTCAG and 5’- CTGAATCTGTGAGAGCTGCACCGCGTAGCGACCTTCTGTTTCTGC. Mutagenesis was performed according to manufacturer protocol. DNA sequencing verified the C401A mutation was incorporated into K10C401A-2B (Keck Biotechnology Resource Laboratory, Yale University).
Protein Production and Purification
K1-2B was co-expressed in Escherichia coli BL21 (DE3) cells with either K10-2B or K10C401A-2B according to established protocols [14]. Protein isolation, protein purification, and His6-tag removal by thrombin cleavage were also performed as previously reported [14]. The final K1/K10-2B and K1/K10C401A-2B complexes were in 0.1 M Tris-HCl buffer (pH 7.5) containing 0.2 M NaCl and 10 mM 2-mercaptoethanol.
Multi-angle Light Scattering
K1/K10-2B and K1/K10C401A-2B (~ 2.5 mg/ml) in 20 mM Tris-HCl buffer (pH 7.5) containing 0.1 M NaCl were applied separately to a Superdex 75 gel filtration column in-line with a DAWN HELEOS II (Wyatt Technology, Santa Barbara, CA) light scattering instrument at 0.5 ml/min. Laser wavelength was 658 nm. Data collection and analysis used Astra software version 6.1.6.5 (Wyatt Technology).
Crystallization and X-ray Data Collection
The K1/K10C401A-2B heterocomplex at 8.0 mg/ml in 100 mM Tris-HCl buffer (pH 7.5) containing 0.2 M NaCl and 10 mM 2-mercaptoethanol was crystallized by sitting drop vapor diffusion at 12 oC using a reservoir solution of 100 mM Tris-HCl (pH 7.4) containing 1.5 M ammonium phosphate dibasic and 1% N-Octyl-β-D-glucoside. Crystallization drops were prepared by mixing 2 µl protein with 2 µl reservoir solution. Crystals were soaked for ~2 min in a cryoprotectant solution containing 20% glycerol in mother liquor before flash freezing in liquid nitrogen. A native dataset was collected on a single crystal maintained at ~100 K and λ = 0.9791 Å using NE-CAT beamline 24-ID-C at the Advanced Photon Source at Argonne National Laboratory. Diffraction data were processed using HKL-2000 [27].
Structure Determination, Refinement, and Analysis
The human K1/K10C401A-2B structure was determined by molecular replacement using MOLREP [28] and the native K1/K10-2B structure as the initial search model (PDB ID 4ZRY). Molecular replacement bias was reduced by using a simulated annealing composite omit map generated in PHENIX [29]. The K1/K10C401A-2B structure underwent iterative rounds of model building using Coot [30] and refinement using PHENIX with standard geometric and secondary structure restraints. The crystal asymmetric unit contained one K1/K10C401A-2B dimer. The final Ramachandran statistics were: residues in favorable regions, 99%; in allowed regions, 1% (Table 2). The final Rfree value (27.7%) is consistent with other large coiled-coil structures [31]. UCSF Chimera (Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco) and Coot were used to perform the structural analyses. Figures were prepared using UCSF Chimera and Adobe Illustrator.
Table 2.
Data collection and refinement statistics.
| Crystal | K1-K10C401A Heterodimer |
| Diffraction Data* | |
| Space Group | P 61 2 2 |
| Unit Cell Dimensions, a, b, c (Å) α, β, γ (°) |
100.12, 100.12, 224.79 90, 90, 120 |
| Resolution range (outer shell), Å | 48.86-2.07 (2.12-2.07)† |
| I/σI | 22.6 (0.5) |
| CC(1/2) in outer shell, % | 10.7 |
| Completeness, % | 99.4 (98.6) |
| Rmeas | 0.116 (6.606) |
| No. of crystals used | 1 |
| No. of unique reflections | 41,346 |
| Redundancy | 10.5 (10.7) |
| Wilson B-factor, Å2 | 56.1 |
| Refinement | |
| Rwork, % | 0.250 (0.46) |
| Rfree, % | 0.277 (0.50) |
| No. of Non-Hydrogen Atoms | |
| Protein | 1753 |
| Ligands/Ions | 26 |
| Waters | 107 |
| R.m.s. Deviations | |
| Bond lengths, (Å) | 0.004 |
| Angles, (o) | 0.611 |
| Chirality | 0.029 |
| Planarity | 0.002 |
| Dihedral, (o) | 18.546 |
| Average B-factor (overall), Å2 | 123.0 |
* Data collection performed on 04-07-2017. † Values in parentheses are for highest-resolution (outer) shell.
Data Availability
Atomic coordinates and structure factors are deposited in the Protein Data Bank under accession code 6UUI.
Results
K1/K10C401A-2B is a dimer in solution
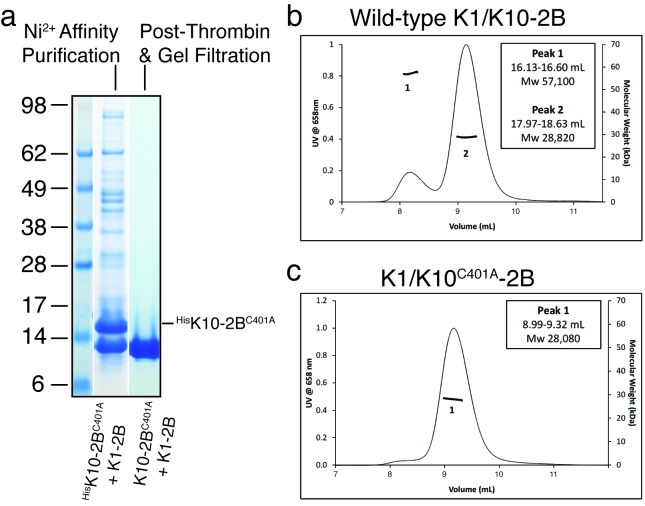
To determine whether the K10C401A mutation altered the ability of K10-2B to bind to K1-2B, we co-expressed His6-K10C401A-2B with untagged K1-2B and subjected the cellular lysate to nickel-affinity purification. His6-K10C401A-2B bound K1-2B in a 1:1 stoichiometry, which enabled purification of the K1/K10C401A-2B heterocomplex (Figure 3a). After His6-tag removal, K1/K10C401A-2B was analyzed by multi-angle light scattering (MALS) to determine the oligomeric state of the complex compared to the wild-type K1/K10-2B complex (Figure 3b-c). Native K1/K10-2B was predominantly a heterodimer in solution (Peak 2 observed MW 28,820; calculated MW 27,899); however, a small fraction of a tetrameric species was observed (Peak 1 observed MW 57,100; calculated MW 55,798) (Figure 3b). In contrast, MALS of K1/K10C401A-2B produced a single peak corresponding to a heterodimer (observed MW 28,080) without evidence of a tetrameric species (Figure 3c). This data supports the conclusion that the tetrameric species in solution for native K1/K10-2B occurs due to K10C401-mediated disulfide linkage and that the C401A mutation indeed abolishes disulfide bonding capability at position 401 on K10. This is in agreement with our prior biochemical analysis showing the reducing agent TCEP can abrogate K10C401-mediated dimerization on a non-reducing denaturing gel [14].
Figure 3.
Light scattering demonstrates K1/K10C401A-2B is a dimer in solution. (a) HisK10C401A-2B bound untagged K1-2B in the bacterial lysate in a 1:1 stoichiometry and purified as a 1:1 complex using nickel affinity chromatography. After thrombin cleavage of the N-terminal His6 tag, K10C401A-2B migrated on the polyacrylamide gel at the same molecular weight (MW) as K1-2B, as expected. The final purified K1/K10C401A-2B complex after gel filtration is shown (gel lane farthest right). (b) Wild-type K1/K10-2B heterocomplex was primarily a heterodimer in solution (Peak 2), but a small fraction of a tetrameric species was observed (Peak 1) by multi-angle light scattering (MALS). (c) MALS of K1/K10C401A-2B produced a single heterodimer peak without evidence for a tetrameric species, indicating substitution of C401 with alanine abrogates disulfide-mediated tetramer formation.
K1/K10C401A-2B structure
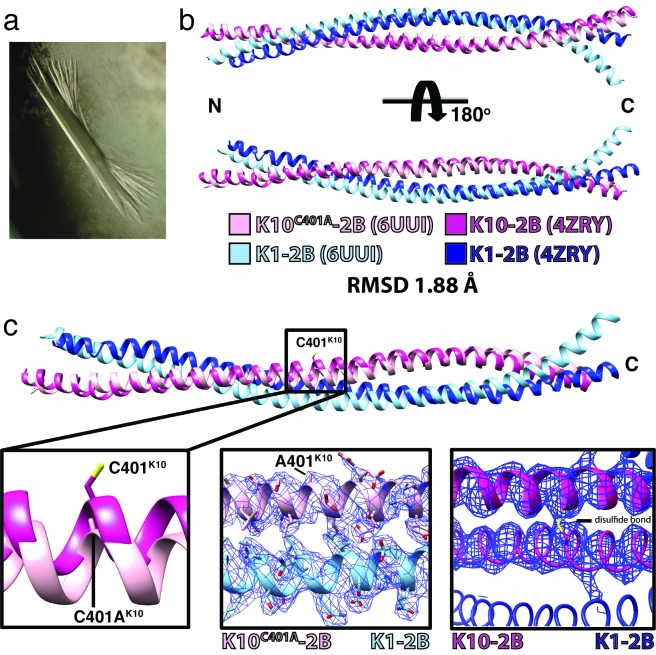
Purified K1/K10C401A-2B, confirmed as a heterodimer in solution by MALS, was crystallized and the X-ray crystal structure of the heterodimeric complex determined at 2.07 Å resolution (Figure 4 and Table 2). The K1 and K10C401A molecules in the heterodimer form a parallel coiled-coil, spanning K1 residues 383-489 and K10 residues 349-456. The N-terminal 13 residues of K1-2B (370-382) and 12 residues of K10-2B (337-348) were flexible, not ordered in the crystal, and unable to be modeled. Superposition of the K1/K10C401A-2B heterodimer structure onto the wild-type K1/K10-2B heterodimer structure had a root-mean-square deviation of 1.88 Å across all atom pairs (Figure 4b). Examination of the superposed structures revealed that the largest difference between the two is enhanced curvature of the K1-2B C-terminus in the K1/K10C401A-2B complex (light blue chain in Figure 4b). The K10-2B molecule containing the C401A mutation has minimal backbone deviation compared to the native structure. Examination of the electron density at position 401 on K10 confirmed the C401A substitution and the absence of a disulfide linkage in the K1/K10C401A-2B structure (Figure 4c).
Figure 4.
Human K1/K10C401A-2B heterodimer crystal structure. (a) Representative crystal of the K1/K10C401A-2B complex. (b) Superposition of the K1/K10C401A-2B heterodimer onto the wild-type K1/K10-2B heterodimer (PDB ID 4ZRY) had a root-mean-square deviation (RMSD) of 1.88 Å across all atom pairs. There is enhanced curvature at the C-terminus of K1-2B in the K1/K10C401A-2B heterodimer (light blue) compared to the wild-type structure. (c) A zoomed in view of the superposed structures from (b) at K10 position 401 demonstrated electron density for an alanine, confirming the K10C401A substitution. There is no structural evidence for a disulfide bond in the K1/K10C401A-2B heterodimer.
Crystal lattice packing of K1/K10C401A-2B
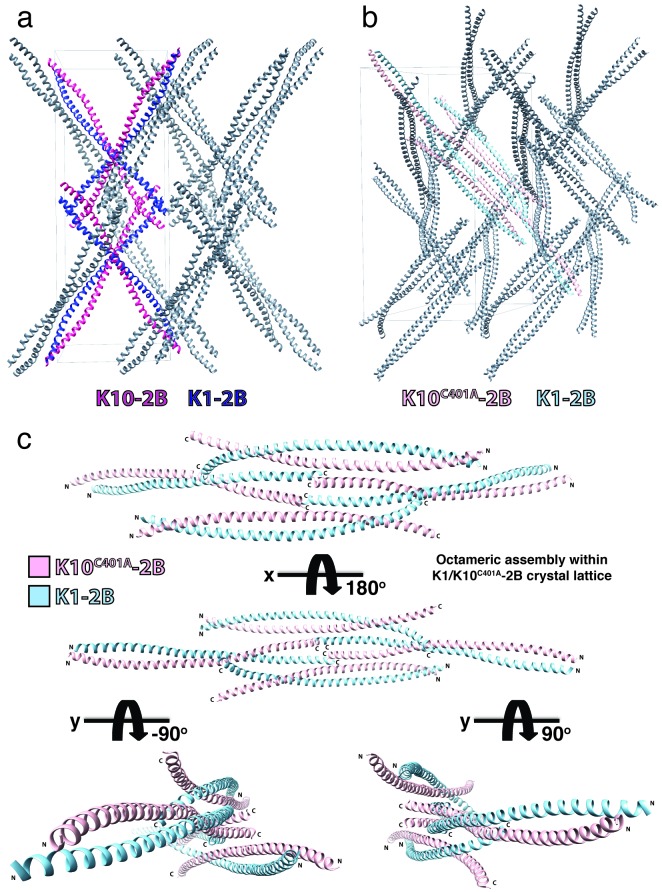
Surprisingly, despite achieving high resolution diffraction, the estimated solvent content of the K1/K10C401A-2B crystals is still ~ 79% with a Matthews coefficient of 5.85. The K10C401A mutation caused a different packing of molecules in the crystal lattice compared to the one of wild-type K1/K10-2B. No disulfide-linked X-shape structures are present in the K1/K10C401A-2B crystal lattice (Figure 5). Within the K1/K10C401A-2B crystal lattice there exists an octameric assembly of 2B heterodimers (Figure 5c). This octameric assembly consists of two major heterodimer interactions: 1) an approximately half-staggered anti-parallel alignment of 2B heterodimers and 2) a splayed C-terminal tetrameric interface.
Figure 5.
Comparison of the crystal lattice packing for wild-type K1/K10-2B and K1/K10C401A-2B structures. (a) The crystal lattice for the wild-type K1/K10-2B heterodimer (PDB ID 4ZRY) demonstrated an “X-shaped” structure due to a transdimer disulfide bond between K10C401 residues on adjacent heterodimers. Two examples of the X-shaped tetrameric species are colored (K1-2B, dark blue; K10-2B, magenta). (b) The crystal lattice for K1/K10C401A-2B did not contain a disulfide-linked X-shaped structure. It did contain an octameric assembly, which is colored (K1-2B, light blue; K10C401A-2B, pink). (c) The octameric K1/K10C401A-2B assembly from the crystal lattice is shown separately for clearer viewing; its contacts are mediated largely by the C-terminal aspect of helix 2B.
Key amino acid interactions in K1/K10C401A-2B octameric assembly
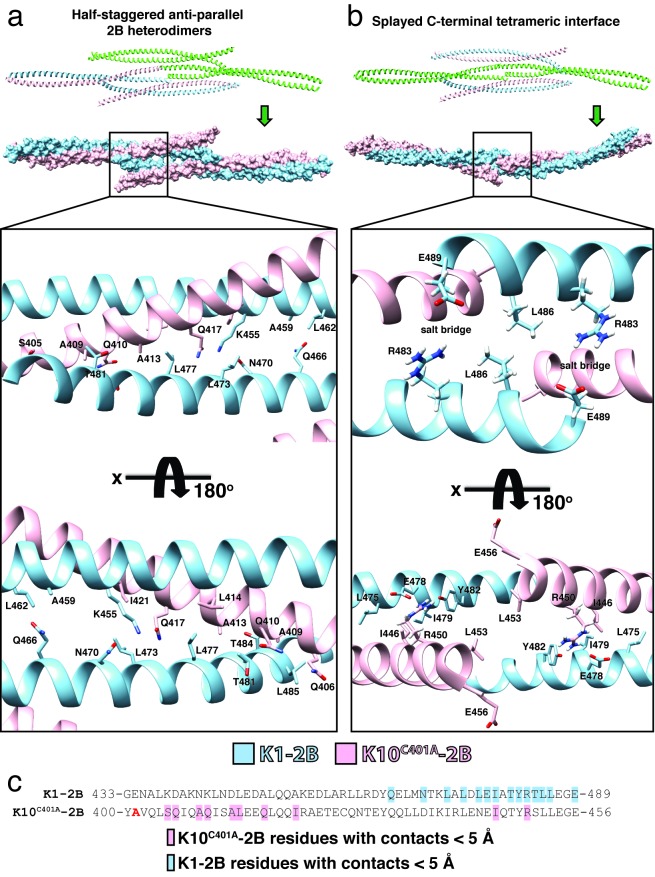
The molecular interactions driving the half-staggered anti-parallel 2B heterodimers and the splayed C-terminal tetrameric interface involve a discrete set of residues: notably, the C-terminus of K1-2B plays a major role in both complexes (Figure 6). In the half-staggered anti-parallel 2B heterodimers there are five K1-2B residues (L473, L477, T481, T484, L485) that interact with eight K10C401A-2B residues (S405, Q406, A409, Q410, A413, L414, Q417, I421) over distances less than 5 Å (Figure 6a; Table 3). The half-staggered anti-parallel 2B complex also involves four K1-K1’ interactions: Q466-A459’, Q466-L462’, N470-K455’, and L473-K455’. All of these interactions occur twice in a single half-staggered anti-parallel 2B complex due to an axis of symmetry at R463K1.
Figure 6.
Residue interactions contributing to the K1/K10C401A-2B octameric assembly. (a) Within the K1/K10C401A-2B octameric assembly, one tetrameric interface is defined by half-staggered anti-parallel 2B heterodimers (top, green). The molecular surface for the half-staggered anti-parallel 2B tetramer is shown in pink (K10C401A) and light blue (K1-2B). Specific amino acid interactions by the C-terminus of K1-2B that are critical for tetramer stabilization are depicted (lower half). (b) A second tetrameric interface within the K1/K10C401A-2B octameric assembly occurs exclusively between the splayed C-terminal ends of two 2B heterodimers: hence, a “splayed C-terminal tetrameric interface” (top, green). Its molecular surface is shown in pink (K10C401A) and light blue (K1-2B). Specific amino acid interactions by the C-termini of K1-2B and K10C401A that are critical for tetramer stabilization are depicted, including important salt bridges (lower half). (c) Partial, aligned, amino acid sequences for the C-terminal half of helix 2B for K1 and K10C401A. Residues participating in contacts < 5 Å in either the half-staggered or splayed tetrameric interface are colored light blue (K1) or pink (K10C401A) on the sequence. There is a clustering of residues involved in important contacts at the C-terminus of K1-2B and the K10-2B region just distal to the C401A substitution site (bold red “A”).
Table 3. Specific residue interactions and distances in K1/K10C401A-2B octameric assembly. All contact distances reported are based on molecular distances between residues that are < 5 Å and were calculated without hydrogens atoms modeled except for where denoted with an asterisk (*). Distances involving residues with salt bridge formation were calculated with hydrogens modeled (*). Salt bridge interactions generally occur over distances < 4 Å.
| Half-staggered anti-parallel 2B heterodimers | Splayed C-terminal tetrameric interface | |||
| K1-2B − K10C401A-2B Interactions (distance, Å) | K1-2B − K1’-2B Interactions (distance, Å) | K1-2B − K1’-2B Interactions (distance, Å) | K1-2B − K10C401A-2B Interactions (distance, Å) | |
| L473−I421 (4.44) | T481−Q410 (4.27) | Q466−A459’ (3.99) | R483−E489’ (4.09/3.52*) | L475−I446 (4.52) |
| L473−Q417 (3.80) | T481−A409 (4.41) | Q466−L462’ (4.17) | L486−L486’ (3.80) | E478−R450 (4.38/3.65*) |
| L477−Q417 (3.69) | T484−A409 (3.99) | N470−K455’ (2.88) | E489−R483’ (4.25/3.52*) | I479−I446 (3.36) |
| L477−L414 (3.90) | L485−A409 (4.00) | L473−K455’ (4.84) | Y482−R450 (4.00/3.51*) | |
| L477−A413 (3.73) | L485−Q406 (4.99) | |||
| T481−A413 (3.45) | L485−S405 (4.40) | |||
K1 residues are also critical to the stabilization of the splayed C-terminal tetrameric interface (Figure 6b). This complex is primarily stabilized by symmetric salt bridges from K1 residues R483 and E489; they surround a hydrophobic interaction between L486K1 from partner helices. Other contributory interactions to the splayed C-terminal tetrameric interface involve four K1-2B residues (L475, E478, I479, Y482) and two K10C401A-2B residues (I446, R450). R450K10 is involved in an intra-heterodimer salt bridge with E478K1 and Y482K1.
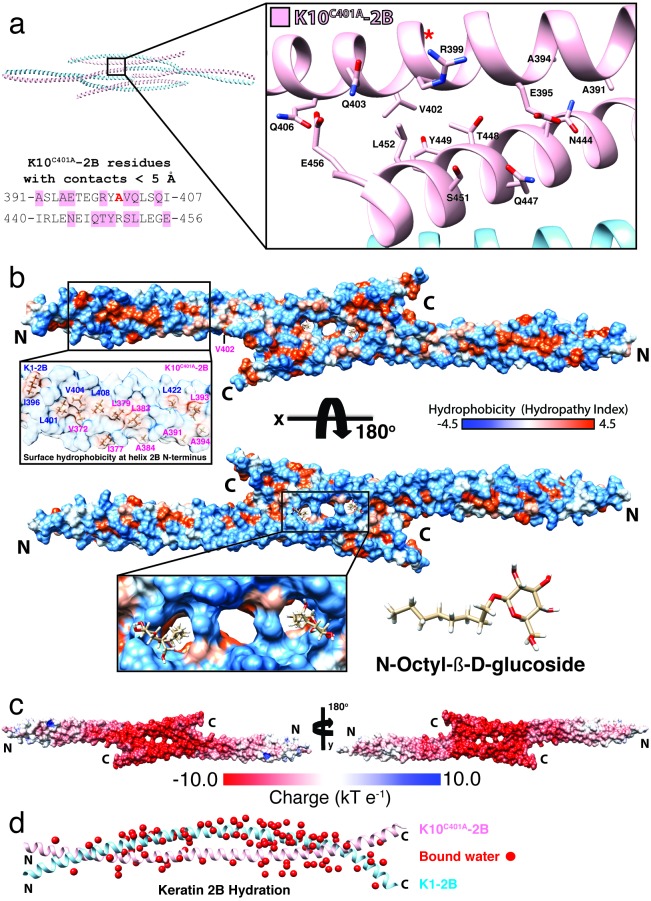
Two adjacent half-staggered anti-parallel 2B heterodimers have one additional K10-K10’ contact interface that helps stabilize the octameric assembly (Figure 7a). This interface consists of seven K10C401A-2B residues (A391, A394, E395, R399, V402, Q403, Q406) around the C401A site that contact seven C-terminal K10C401A-2B’ residues (N444, Q447, T448, Y449, S451, L452, E456) over distances less than 5 Å.
Figure 7.
Molecular surface analysis of K1/K10C401A-2B half-staggered tetramer. (a) Within the K1/K10C401A-2B octameric assembly, two half-staggered anti-parallel tetramers make a contact at a K10-K10’ interface. The K10 residues involved in this interface are depicted structurally (inset, pink) and mapped onto the amino acid sequence (left). One K10 interface (391-407) occurs around the C401A site (red “A”; “*” denotes the structural location of K10C401A whose side chain faces away from the viewer). The other K10 interface (440-456) occurs at the C-terminus of K10-2B. (b) Molecular surface of the K1/K10C401A-2B half-staggered anti-parallel tetramer colored by hydrophobic potential (orange is more hydrophobic). Surface-exposed hydrophobic residues are clustered at the N-terminus of the 2B heterodimer (inset, left). Other surface-exposed hydrophobic residues identified in the previous wild-type structure are buried in the symmetric C-terminal binding interface, which contains three adjacent cavities in the center of the tetramer (panel b, bottom). Two N-Octyl-β-D-glucoside molecules are bound in these cavities. (c) Molecular surface of the K1/K10C401A-2B half-staggered anti-parallel tetramer colored for electrostatic potential (red, acidic; blue, basic). The symmetric C-terminal binding interface at the center of the half-staggered anti-parallel tetramer is highly acidic. (d) 107 bound waters (red spheres) were modeled in the K1/K10C401A-2B heterodimer structure.
Molecular surface features of the K1/K10C401A-2B half-staggered tetramer
Hydrophobic surface potential was mapped onto the molecular surface of the half-staggered anti-parallel tetramer and demonstrated nearly identical solvent-accessible hydrophobicity at the N-terminal half of the K1/K10C401A-2B heterodimer as the prior wild-type structure (Fig 7b) [14]. Five K1-2B residues (I396, L401, V404, L408, L422) and eight K10C401A-2B residues (V372, I377, L379, L383, A384, A391, L393, A394) contribute to solvent-accessible hydrophobic surface at the N-terminus of the 2B heterodimer (inset, Figure 7b). Two surface-exposed leucine residues that were prominent at the C-terminus of the wild-type K1/K10-2B structure (L473K1, L477K1) are buried in the contact interface of the half-staggered anti-parallel tetramer (Figure 6a). Two other hydrophobic residues (L436K10, I440K10) that were surface-exposed in the wild-type structure remain surface-exposed in the K1/K10C401A-2B structure. L452K10, which was surface-exposed in the wild-type structure, participates in the K10-K10’ contact interface in the octameric assembly (Figure 7a).
Crystal size and morphology was improved with the addition of N-Octyl-β-D-glucoside to the crystallization solution; therefore, it was not surprising to identify two bound N-Octyl-β-D-glucoside molecules in the structure (Figure 7b, lower inset). They bound at a symmetric central cavity created by a gap in the contact interface of the half-staggered anti-parallel tetramer. Besides binding N-Octyl-β-D-glucoside, this particular contact interface is highly acidic. Mapping of electrostatic surface potential onto the molecular surface of K1/K10C401A-2B in the context of the half-staggered anti-parallel tetramer revealed that the contact interface driving tetramer formation is highly acidic (the most acidic region of the entire structure) (Figure 7c). One-hundred-and-seven bound waters were identified in the K1/K10C401A-2B structure (Figure 7d).
K1/K10C401A-2B structure does not capture A22 tetrameric alignment
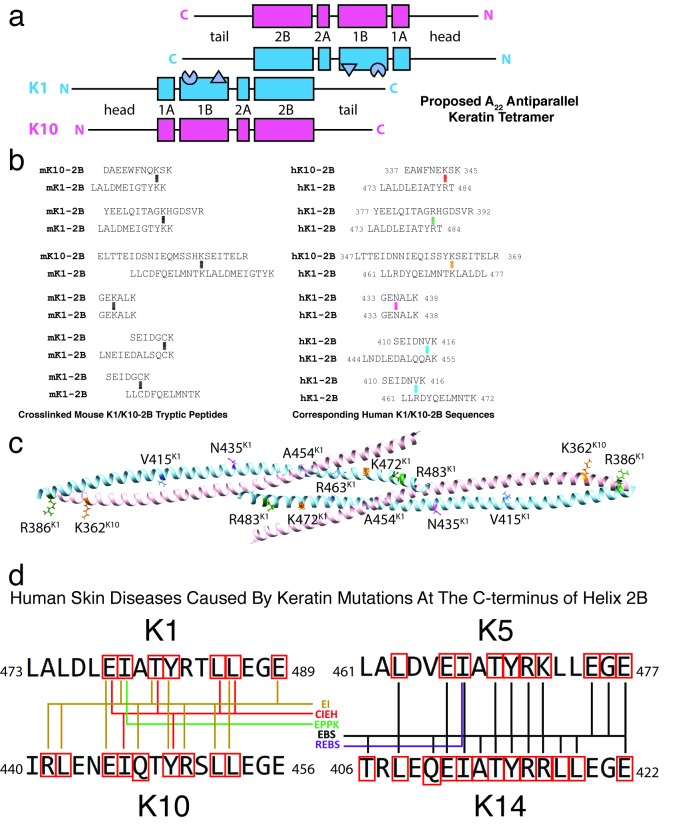
The A22 mode of tetramer alignment as proposed by Steinert and colleagues from cross-linking studies [13,32] describes two anti-parallel keratin heterodimers whose 2B domains are in-phase (not half-staggered) and planar (the interaction is not at an angle) (Figure 8a). Analysis of the unit cell of the K1/K10C401A-2B structure did not show any 2B heterodimers packed in-phase (Figure 5), but it did show a half-staggered anti-parallel tetramer (Figures 5-7). To fully evaluate whether any of the previously identified chemical cross-links establishing the A22 mode were present in the K1/K10C401A-2B half-staggered anti-parallel tetramer structure, we translated the cross-link sequences from mouse to human and mapped them onto the K1/K10C401A-2B structure (Figures 8b,c). Of the six pairs of cross-links (involving 8 lysines and 4 cysteines), only three of the lysine residues are conserved in humans and none of the cysteine residues. Three other lysine residues are arginine residues in the human sequence. In total, none of the corresponding human residues displayed the expected proximity for an A22 alignment based on the previous cross-linking experiments [13].
Figure 8.
The K1/K10C401A-2B structure does not capture the A22 tetrameric alignment. (a) Schematic of the proposed A22 tetramer alignment based on prior cross-linking and nearest neighbor analysis. The two K1/K10 heterodimers have their 2B regions in-phase. The knob (triangle) and pocket (partial circle) structural elements identified in the 1B region are depicted. (b) Six cross-linked mouse K1- and mouse K10-2B tryptic peptides were identified in prior biochemical cross-linking studies on mature K1/K10 IFs (left). The mouse peptide sequences are translated into their corresponding human sequences (right), demonstrating many of the lysines involved in the mouse keratin cross-links are not conserved in humans. (c) The human residues corresponding to the key lysine residues involved in mouse cross-links are mapped onto the K1/K10C401A-2B half-staggered anti-parallel tetramer structure. (d) The amino acid sequence for seventeen residues at the C-terminus of helix 2B for K1/K10 and K5/K14 are illustrated. Five human skin diseases map to mutations in this “mutational hotspot” in keratins 1/10 and 5/14: epidermolytic ichthyosis (EI; formerly known as epidermolytic hyperkeratosis or bullous congenital ichthyosiform erythroderma), cyclic ichthyosis with epidermolytic hyperkeratosis (CIEH), epidermolytic palmoplantar keratoderma (EPPK), epidermolysis bullosa simplex (EBS, including Koebner, Weber-Cockayne, and Dowling-Meara types), and recessive epidermolysis bullosa simplex (REBS). Residues that are mutated in one of these skin diseases are enclosed in a red box. Colored lines connect the residue with the appropriate disease listed in the center based upon keratin mutations documented online in the Human Intermediate Filament Database.
Discussion
There are 54 different keratin proteins, but only four have experimentally determined structures (K1/K10 and K5/14 complexes, Table 1). This structural knowledge gap for keratins is important to overcome because keratins are significant players in human health and disease [1,3]. It is well-established that a “divide-and-conquer” approach to crystallization of IF proteins is beneficial because it reduces the size and flexibility of the crystallization target [31,33]. However, in the wild-type K1/K10-2B crystal structure, even the smaller 2B fragment demonstrated enhanced flexibility at the N-terminus of K10 and achieved modest resolution (3.3 Å) [14]. We hypothesized that the resolution for that structure was limited due to a transdimer disulfide bond between K10C401 residues which generated an X-shaped structure (Figure 1d). Here, we substituted the cysteine with alanine and observed several outcomes in the human K1/K10C401A-2B structure: 1) the disulfide bond was eliminated, 2) the crystal lattice packing was altered, and 3) the diffraction data obtained is of the highest resolution to date for any keratin structure (Table 1).
Interestingly, despite the C401A substitution occurring in K10, it was K1-2B that demonstrated the largest helical deviation when the K1/K10C401A-2B structure was superposed onto the wild-type K1/K10-2B structure (Figure 4b). This occurred because the K1-2B C-terminus contributed several molecular contacts in the crystal lattice that stabilized an octameric assembly, composed of what we termed a half-staggered anti-parallel tetramer and a splayed C-terminal tetrameric interface (Figures 5 and 6). We hypothesized that the C401A substitution would enable the K1/K10C401A-2B heterodimer to pack in the A22 tetramer mode of alignment [13,34]; however, biophysical analysis using MALS demonstrated K1/K10C401A-2B is a heterodimer in solution. This is in contrast to the K1/K10-1B A11 tetramer structure, where prior to crystallization a stable tetrameric complex was observed in solution [19]. It is critical that solution biophysics, in vitro filament assembly, or cell biology data exist to support conclusions made about IF assembly from crystal lattice contacts. Our analysis of the K1/K10C401A-2B half-staggered anti-parallel tetramer observed in the crystal lattice did not recapitulate any of the keratin cross-links identified by Steinert and colleagues in mice (Figure 8). Therefore, we conclude that the octameric assembly observed here is likely not a physiological complex present within a mature 10-nm IF.
While it is important to not over-interpret the physiological relevance of crystal lattice contacts in a crystal, some of the molecular contacts in the octameric assembly may reflect potential interactions in K1/K10 IFs at the reducing conditions in the cell. We observed multiple molecular contacts in the C-terminus of K1/K10C401A-2B, a region known to be a mutational hotspot for clinically-relevant human skin diseases (Figure 8d). Furthermore, it has been shown that the “R/KLLEGE” sequences in the K5 and K14 2B C-terminus regulate lateral alignment within K5/K14 IFs, not filament elongation [22]. Additionally, there are surface-exposed C-terminal hydrophobic residues in the wild-type K1/K10-2B structure (e.g. L473K1, L477K1) that are buried in the contact interface of the half-staggered anti-parallel tetramer, suggesting surface-exposed hydrophobic residues at the C-terminus of helix 2B may play a role in lateral assembly of IFs. Lastly, the helix 2B electrostatic potential at the C-terminus is very acidic–the most acidic aspect of the molecule. In total, there are four lines of evidence that the C-terminal region of helix 2B plays an important role in IF assembly–likely the lateral assembly of protofilaments and/or protofibrils: 1) molecular contacts in this crystal structure, 2) pathologic mutations from patients, 3) in vitro filament data on TYR*LLEGE sequences conserved in keratins, 4) distinct hydrophobic and acidic molecular surface properties. We interpret this collection of data to mean that the helix 2B C-terminus potentially functions as a regulator of lateral packing within a mature 10-nm filament (but not necessarily the only regulator). This is consistent with our recent model for IF assembly that centers around the A11 tetramer as the building block for IF assembly based on the biochemical and structural evidence available [19].
It is important to strive for atomic resolution structures that more clearly delineate how specific IF regions regulate length and width during IF assembly. This level of molecular detail is essential to understanding the genotype-structurotype-phenotype correlation for IF-related diseases. It is also a precursor for developing precision therapeutics that target and manipulate the IF networks in cells.
Acknowledgments
We thank the late Prof. Thomas A. Steitz (Dept. of Molecular Biophysics and Biochemistry, Yale Univ.) for providing laboratory space, equipment, and supplies for this project. This work was supported by the Foundation for Ichthyosis & Related Skin Types through a Research Award (to CGB) and NIH/NIAMS Award K08-AR070290 (to CGB). Aspects of this work were presented at the 2018 International Investigative Dermatology Meeting (Orlando) [35].
Glossary
- IF
intermediate filament
- PDB
Protein Data Bank
- KIF
keratin intermediate filament
- GFAP
glial fibrillary acidic protein
- EM
electron microscopy
- MALS
multi-angle light scattering
- MW
molecular weight
Author Contributions
CGB designed the study. IBL conducted the mutagenesis, crystallization, and x-ray data collection. IBL and CGB processed and refined the crystallographic data, and performed the structural analyses with assistance from AJH. AJH performed light scattering. CGB wrote the manuscript with assistance from ILB, AJH, SE, and MH. All authors approved the final manuscript.
References
- Omary MB. “IF-pathies”: a broad spectrum of intermediate filament-associated diseases. J Clin Invest. 2009;119(7):1756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bär H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8(7):562–73. [DOI] [PubMed] [Google Scholar]
- Toivola DM, Boor P, Alam C, Strnad P. Keratins in health and disease. Curr Opin Cell Biol. 2015;32:73–81. [DOI] [PubMed] [Google Scholar]
- Toivola DM, Strnad P, Habtezion A, Omary MB. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20(2):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Strnad P, Hanada S. Toward unraveling the complexity of simple epithelial keratins in human disease. J Clin Invest. 2009;119(7):1794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YW, Runyan RB, Oshima RG, Hendrix MJ. Expression of complete keratin filaments in mouse L cells augments cell migration and invasion. Proc Natl Acad Sci USA. 1993;90(9):4261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YW, Seftor EA, Romer LH, Hendrix MJ. Experimental coexpression of vimentin and keratin intermediate filaments in human melanoma cells augments motility. Am J Pathol. 1996;148(1):63–9. [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Mack JW, Korge BP, Gan SQ, Haynes SR, Steven AC. Glycine loops in proteins: their occurrence in certain intermediate filament chains, loricrins and single-stranded RNA binding proteins. Int J Biol Macromol. 1991;13(3):130–9. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci. 2006;31(7):383–94. [DOI] [PubMed] [Google Scholar]
- Badowski C, Sim AY, Verma C, Szeverényi I, Natesavelalar C, Terron-Kwiatkowski A, et al. Modeling the Structure of Keratin 1 and 10 Terminal Domains and their Misassembly in Keratoderma. J Invest Dermatol. 2017;137(9):1914–23. [DOI] [PubMed] [Google Scholar]
- Nicolet S, Herrmann H, Aebi U, Strelkov SV. Atomic structure of vimentin coil 2. J Struct Biol. 2010;170(2):369–76. [DOI] [PubMed] [Google Scholar]
- Parry DA. Hendecad repeat in segment 2A and linker L2 of intermediate filament chains implies the possibility of a right-handed coiled-coil structure. J Struct Biol. 2006;155(2):370–4. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Fraser RD, Parry DA. Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J Mol Biol. 1993;230(2):436–52. [DOI] [PubMed] [Google Scholar]
- Bunick CG, Milstone LM. The X-Ray Crystal Structure of the Keratin 1-Keratin 10 Helix 2B Heterodimer Reveals Molecular Surface Properties and Biochemical Insights into Human Skin Disease. J Invest Dermatol. 2017;137(1):142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Marekov LN, Steinert PM. Subfilamentous protofibril structures in fibrous proteins: cross-linking evidence for protofibrils in intermediate filaments. J Biol Chem. 2001;276(42):39253–8. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate Filaments: structure and Assembly. Cold Spring Harb Perspect Biol. 2016;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Fowler WE, Rew P, Sun TT. The fibrillar substructure of keratin filaments unraveled. J Cell Biol. 1983;97(4):1131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Groppi VE, Gui H, Chen L, Xie Q, Liu L, et al. High-Throughput Screening for Drugs that Modulate Intermediate Filament Proteins. Methods Enzymol. 2016;568:163–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldirany SA, Ho M, Hinbest AJ, Lomakin IB, Bunick CG. Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J. 2019;38(11):e100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers A, Kuhn A, Mahrle G. Palmoplantar keratoderma with tonotubular keratin. J Am Acad Dermatol. 1991;24(4):638–42. [DOI] [PubMed] [Google Scholar]
- Terron-Kwiatkowski A, van Steensel MA, van Geel M, Lane EB, McLean WH, Steijlen PM. Mutation S233L in the 1B domain of keratin 1 causes epidermolytic palmoplantar keratoderma with “tonotubular” keratin. J Invest Dermatol. 2006;126(3):607–13. [DOI] [PubMed] [Google Scholar]
- Wilson AK, Coulombe PA, Fuchs E. The roles of K5 and K14 head, tail, and R/K L L E G E domains in keratin filament assembly in vitro. J Cell Biol. 1992;119(2):401–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol. 2012;19(7):707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick CG. 349 X-ray crystal structure of the keratin 1-keratin 10 heterodimer reveals a molecular basis for associated keratinopathies. J Invest Dermatol. 2015;135 Suppl 1:S58–69.25865012 [Google Scholar]
- Feng X, Coulombe PA. A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J Cell Biol. 2015;209(1):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Parry DA. Conservation of the structure of keratin intermediate filaments: molecular mechanism by which different keratin molecules integrate into preexisting keratin intermediate filaments during differentiation. Biochemistry. 1993;32(38):10046–56. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: C.W. Carter JRMS, editor. Methods in Enzymology. Macromolecular Crystallography, part A. 276. New York: Academic Press; 1997. p. 307-26. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–5. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126-32. [DOI] [PubMed] [Google Scholar]
- Chernyatina AA, Hess JF, Guzenko D, Voss JC, Strelkov SV. How to Study Intermediate Filaments in Atomic Detail. Methods Enzymol. 2016;568:3–33. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Parry DA. Conservation of the structure of keratin intermediate filaments: molecular mechanism by which different keratin molecules integrate into preexisting keratin intermediate filaments during differentiation. Biochemistry. 1993;32(38):10046–56. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, et al. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J Mol Biol. 2001;306(4):773–81. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Parry DA. The conserved H1 domain of the type II keratin 1 chain plays an essential role in the alignment of nearest neighbor molecules in mouse and human keratin 1/keratin 10 intermediate filaments at the two- to four-molecule level of structure. J Biol Chem. 1993;268(4):2878–87. [PubMed] [Google Scholar]
- Lomakin IB, Hinbest A, Bunick CG. 713 The crystal structure of keratin 1/10(Cys401Ala) helix 2B heterodimer determined at 2.0 Å resolution. J Invest Dermatol. 2018;138(5):S121. [Google Scholar]
- Lee C, Kim M, Li S, Leahy D, Coulombe P. Structure-function analyses of a keratin heterotypic complex identify specific keratin regions involved in intermediate filament assembly. bioRxiv. 2019:821090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates and structure factors are deposited in the Protein Data Bank under accession code 6UUI.