Abstract
Porcine deltacoronavirus (PDCoV) is a novel coronavirus that can cause vomiting and watery diarrhea in pigs and death in piglets. Since PDCoV was first detected in 2009 in Hong Kong, the prevalence of PDCoV has increased in recent years, resulting in serious economic losses to the swine industry. The coronavirus spike (S) protein is an antigen that has been demonstrated to contain epitopes that induce neutralizing antibodies. The presence of serum and milk IgA antibodies against pathogens that replicate primarily on mucosal surfaces is important for mucosal immunity. Here, an indirect anti-PDCoV IgA antibody enzyme-linked immunosorbent assay (PDCoV S1 IgA ELISA) using the purified S1 portion of S protein as the coating antigen was developed to detect PDCoV IgA antibodies in serum and sow’s milk. A receiver operating characteristic (ROC) curve analysis showed high specificity and sensitivity of the PDCoV-S1-IgA-ELISA based on samples confirmed by IFA. Anti-PDCoV IgA antibodies in 152 serum samples and 65 milk samples collected from six farms that had experienced diarrhea outbreaks within previous last two years were detected by this assay, and 62.5% of the serum samples and 100% of the milk samples were positive for PDCoV. The indirect ELISA method established in this study will provide a convenient tool for measurement of serum and milk IgA levels against PDCoV in pig herds, rapid detection of PDCoV infection in pigs, and evaluation of the immunogenicity of vaccines.
Electronic supplementary material
The online version of this article (10.1007/s00705-020-04541-6) contains supplementary material, which is available to authorized users.
Introduction
Porcine deltacoronavirus (PDCoV) is a novel coronavirus that was first discovered in 2009 in Hong Kong [1]. The virus was then identified in the United States in February 2014, and severe outbreaks soon occurred in multiple states in the United States [2–5]. Shortly thereafter, PDCoV was identified in South Korea, mainland China, Thailand, Canada, and Laos [6–9]. Since PDCoV was detected in mainland China in 2015, the prevalence of PDCoV has continued to increase and has resulted in serious economic losses to the swine industry [10, 11]. PDCoV strain NH was successfully isolated from the small intestine of sick piglets using porcine kidney cells in China [12].
PDCoV is an enveloped, single-stranded, positive-sense RNA virus with a genome length of appropriately 25 kb belonging to the genus Deltacoronavirus, family Coronaviridae [13]. Like other coronaviruses, PDCoV also contains four main structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The S protein is postulated to contain epitopes that induce neutralizing antibodies, and it has receptor-binding, cell membrane fusion, and virus entry functions [14, 15]. In most coronaviruses, the S protein is cleaved into two separate polypeptides, S1 and S2, by a host cell furin-like protease. S1 is the large receptor-binding domain, while S2 mediates membrane fusion and virus entry [16–18]. Enzyme-linked immunosorbent assays (ELISAs) for the detection of antibodies against porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV) based on the purified S1 portion of the S protein have been successfully developed [19–21].
Several reports have described ELISAs for detection of neutralizing antibodies or immunoglobulin (IgG) in serum samples [22–24]. However, PDCoV is an enteric pathogen, and the presence of IgA antibodies against pathogens that replicate primarily on mucosal surfaces is important for mucosal immunity. IgA is an important immunoglobulin for mucosal immunity that can be detected in serum and milk of pigs after virus challenge or inoculation, and therefore serum and milk IgA antibodies to enteric pathogens can act as indicators [25, 26]. Therefore, measuring IgA levels in serum and milk samples is critical for evaluating the level of protection of the mucosal response against PDCoV infection. Here, we have developed an indirect anti-PDCoV IgA antibody ELISA that uses the purified S1 portion of PDCoV S protein as a coating antigen. This assay will serve as a convenient tool for measuring of serum and milk IgA antibody levels in pig herds. Many studies have shown that serum antibodies are passed mainly to piglets through sow’s milk, which results in passive immunoprotection. IgA antibodies have a variety of biological functions, such as inhibiting adhesion and immunological exclusion and neutralization of viruses [27]. Thus, an indirect ELISA for detection of IgA antibodies against PDCoV can be used as a sensitive and specific quantitative method to evaluate immunity to PDCoV.
Materials and methods
Ethics statement
The animal experiments were approved by Harbin Veterinary Research Institute and performed in accordance with animal ethics guidelines and approved protocols. The approval number of the Animal Ethics Committee is Heilongjiang-SYXK-2017-009. All sera and milk samples used in this study were obtained as routine diagnostic samples collected from six pig farms in China with the farm owners’ permission.
Virus, antibodies and cells
PDCoV strain NH and antisera against PDCoV, PEDV, TGEV, porcine rotavirus (PoRV), and porcine circovirus 2 (PCV2) were obtained from the State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences. Monoclonal antibodies against the Fc tag were purchased from Sigma-Aldrich (SAB3701275). Swine testis (ST) and human embryonic kidney 293T (HEK-293T) cells were obtained from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and were cultured under a humidified atmosphere of 5% CO2 at 37 °C.
Synthesis and expression of the PDCoV S1 gene
The region encoding the putative S1 domain (amino acids 1-672) of PDCoV strain NH (GenBank no. KU981062.1) was terminally fused to the Fc domain of human IgG1, synthesized by and purchased from BGI (Beijing, China), and then cloned into the KpnI-EcoRI restriction sites of the eukaryotic expression vector pcDNA3.1(+) to yield the recombinant plasmid pcDNA3.1-S1-Fc. The S1-Fc fusion protein was expressed in 293T cells transfected with pcDNA3.1-S1-Fc, analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and purified using protein A Sepharose CL-4B (GE Healthcare, WI, USA) according to the manufacturer’s instructions.
Western blotting of the recombinant PDCoV S1 protein
Purified PDCoV S1 protein with an Fc-tag was subjected to 12% SDS-PAGE and then transferred to a nitrocellulose (NC) membrane. The NC membrane was blocked using 5% (w/v) nonfat dried milk in phosphate-buffered saline (PBS) at 37 °C for 1 h and then incubated with a rabbit monoclonal antibody (mAb) against the Fc tag (1:20,000) and a porcine polyclonal antibody (pAb) against PDCoV (1:500) at 37 °C for 1 h. After washing three times with PBS containing 0.05% Tween 20 (PBST), the membrane was incubated in PBS containing horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:10,000) (Sigma) or rabbit anti-pig IgG (1:10,000) (Sigma) at 37 °C for 1 h. Membranes were washed with PBST and incubated with substrate solution (PBST, 0.05% DAB and 0.006% H2O2).
Procedure for the PDCoV-S1-IgA-ELISA
ELISA plate wells (Costar, Corning NY, USA) were coated with 0.625 μg of purified PDCoV S1 protein per ml in a bicarbonate/carbonate coating buffer (pH 9.6) at 4 °C overnight. Plates were washed three times with PBST and blocked with 5% (w/v) nonfat dried milk at 37 °C for 2 h. After the plates were washed three times with PBST, the diluted sera or milk (1:100) was added to the wells (100 μl/well) and incubated at 37 °C for 1 h. The plates were washed three times and incubated with 100 μl of HRP-conjugated IgA (1:10,000) (Bethyl Laboratories, Montgomery, TX, USA) at 37 °C for 1 h. After washing three times with PBST, 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) was added to the wells, the plate was incubated for 15 min at room temperature, and the reaction was stopped by adding 50 μl of 2 M H2SO4. OD values were read at 450 nm using an ELx808 Absorbance Microplate Reader (BioTek Instruments, VT, USA).
Determination of the cutoff value for the PDCoV-S1-IgA-ELISA
One hundred twenty-three PDCoV-negative serum samples were tested using the PDCoV S1 IgA ELISA to determine the cutoff value. The reaction conditions were as described above. Each sample was tested three times, and the mean OD450 value was used for analysis. The result for each sample was converted into a percent reactivity (PR) value based on the following formula: PR value = [(OD450 value of the tested sample − OD450 value of the negative control) / (OD450 value of the positive control − OD450 value of the negative control)] × 100%. The PR cutoff value was determined as the mean PR value of the 123 PDCoV-negative sera plus 3 times the standard deviation.
Repeatability test and specificity test
To investigate the reproducibility of the PDCoV S1 IgA ELISA, intra- and inter-assay coefficients of variation (CV) were determined using eight control sera. The intra-assay CV was used to express the reproducibility of the assay within one plate. Eight replicate samples for each concentration were measured to get the intra-assay CV. The inter-assays were carried out on three different days to obtain the inter-assay CV. To evaluate the specificity of this ELISA, antisera against PEDV, TGEV, PoRV, and PCV2 were tested using the PDCoV S1 IgA ELISA. The reaction conditions were the same as those used for the PDCoV S1 IgA ELISA procedures. The PR value of the test samples was calculated. Each sample was tested three times, and the mean PR value was used to determine whether the sample was positive or negative.
Immunofluorescence assay (IFA)
ST cells were seeded in a 96-well tissue culture plate for 48 h. Then, the cell monolayers were washed with PBS and inoculated with 100 median tissue culture infective doses (TCID50) of PDCoV strain NH for 1 h at 37 °C. After removing the inoculum, the cell monolayers were washed three times with PBS and covered with DMEM containing trypsin at a final concentration of 10 μg/ml. At 48 h post-inoculation (hpi), the cells were washed three times with PBS and fixed in 4% paraformaldehyde for 30 min at 4 °C. After washing three times with PBS, the cells were permeabilized with 0.1% Triton X-100 at room temperature for 20 min and then blocked with 5% (w/v) skim milk for 1 h at 37 °C. The cells were incubated with the serum samples (1:100) for 1 h at 37 °C. After washing three times with PBS, the cells were incubated with fluorescein-isothiocyanate-conjugated anti-pig IgA (1:200) (Abcam, Cambridge, MA, USA) or anti-pig IgG (1:500) (Sigma) at 37 °C for 1 h. The cells were then washed three times with PBS and counterstained with DAPI (1:1,000) (Sigma) at room temperature for 20 min to visualize nuclear DNA. After washing with PBS, the specific fluorescence was visualized using an EVOS cell imaging system (Thermo Fisher Scientific, USA).
Validation of the PDCoV-S1-IgA-ELISA
To evaluate the practicality of the PDCoV-S1-IgA-ELISA, a total of 125 serum samples from Hebei Province that had been tested in a neutralization assay (Table S1) were randomly selected. These serum samples were tested using the PDCoV S1 IgA ELISA. Each sample was tested three times. Meanwhile, 125 serum samples were subjected to IFA using PDCoV. The IFA procedure was the same as that described above. To evaluate the sensitivity and specificity of the PDCoV S1 IgA ELISA, a receiver operating characteristic (ROC) curve was generated using the results of the IFA as the standard for negative and positive determination. Statistical analysis was carried out using SPSS software (version 11.5 for Windows, SPSS Inc., Chicago, IL, USA).
Detection of PDCoV in field samples by the PDCoV-S1-IgA-ELISA
A total of 152 serum samples and 65 milk samples were collected from sows from January 2014 to June 2015. The serum samples were collected from Heilongjiang (n = 40), Hebei (n = 40), Beijing (n = 40), and Liaoning (n = 32), and the milk samples were collected from Hefei (n = 46) and Tianjin (n = 19). All serum and milk samples were tested using the PDCoV S1 IgA ELISA. The reaction conditions were the same as those used for the PDCoV S1 IgA ELISA procedures, and each sample was tested three times.
Results
Expression, purification and identification of PDCoV S1 protein
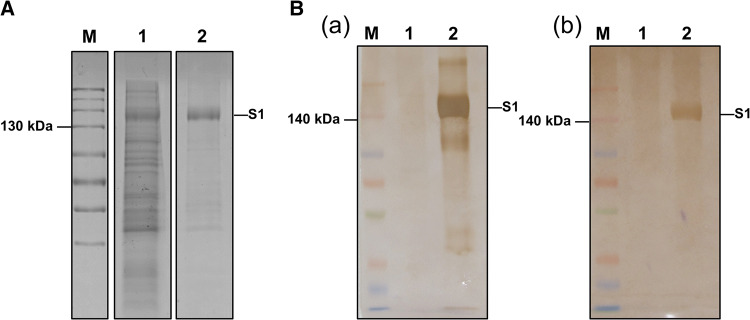
Eukaryotic expression of the synthesized S1 gene of PDCoV was done using the vector pcDNA3.1(+) with an Fc tag. The results showed that the S1 protein of PDCoV was successfully expressed with a Fc tag (Fig. 1A). Furthermore, the purified PDCoV S1 protein was verified by western blotting with anti-Fc-tag mAb and anti-PDCoV pAb, respectively (Fig. 1B).
Fig. 1.
Expression and purification of the PDCoV S1 protein. A. SDS-PAGE analysis of the expression and purification of the PDCoV-S1 protein. Lane M, Spectra™ Multicolor High Range Protein Ladder (26625, 40 kDa-300 kDa); lane 1, the supernatant from the transfected 293T cells; lane 2, purified PDCoV S1 protein (~150 kDa). B. Western blot of the PDCoV-S1 protein with anti-Fc-tag mAb (a) or anti-PDCoV pAb (b). Lane M, Spectra™ Multicolor Broad Range Protein Ladder (10 kDa-260 kDa); lane 1, control; lane 2, purified PDCoV S1 protein (~150 kDa)
Cutoff value of the PDCoV-S1-IgA-ELISA
To determine the cutoff value of the PDCoV S1 IgA ELISA, a total of 123 serum samples were collected from piglets on the farm that was found to be PDCoV negative by RT-PCR (data not shown). These PDCoV-negative serum samples were then tested using the PDCoV S1 IgA ELISA as described above. All OD values were converted to PR values, and a PR cutoff value of 34.51 (mean + 3 × SD, 11.01 + 23.50) was used as the positive standard for the PDCoV S1 IgA ELISA (Fig. 2).
Fig. 2.
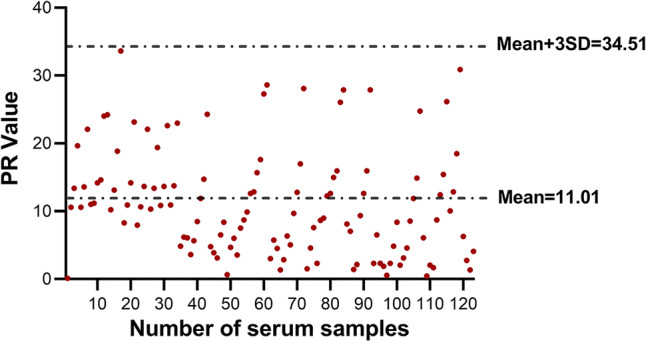
Determination of the cutoff value of the PDCoV S1 IgA ELISA
Repeatability and specificity of the PDCoV S1 IgA ELISA
In the repeat ability experiment, eight control serum samples were used to determine the inter- and intra-assay CV of the PDCoV S1 IgA ELISA, which was 3.12%-8.47% and 1.81%-14.26%, respectively. In the specificity experiment, antisera against PEDV, TGEV, PoRV and PCV2 were used to determine the specificity of the PDCoV S1 IgA ELISA. The results showed that the PR values ranged from 12.11 to 15.07 less than the positive standard, indicating the PDCoV S1 IgA ELISA did not cross-react with these antisera (Fig. 3).
Fig. 3.
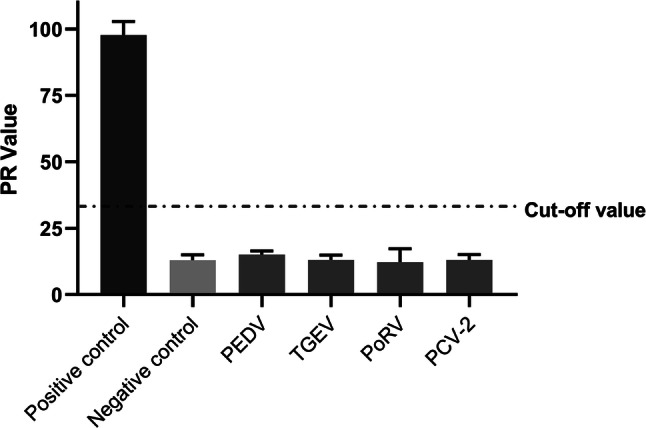
Specificity of the PDCoV S1 IgA ELISA
Validation of the PDCoV S1 IgA ELISA
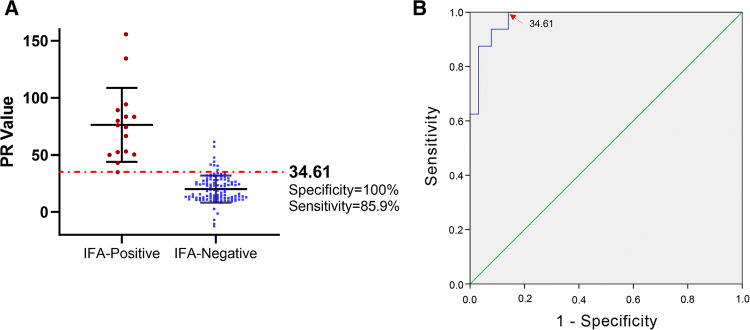
To validate the PDCoV S1 IgA ELISA, a total of 125 serum samples that had been tested by IFA (Fig. 4) were used for ROC analysis. The ROC curve showed that, compared with IFA, the specificity and sensitivity of the PDCoV S1 IgA ELISA was 100% and 85.9%, respectively, when the PR cutoff value was 34.61 (Fig. 5). The optimized PR cutoff value generated from the ROC curve was similar to the PR cutoff value of 34.51 determined using negative sera.
Fig. 4.
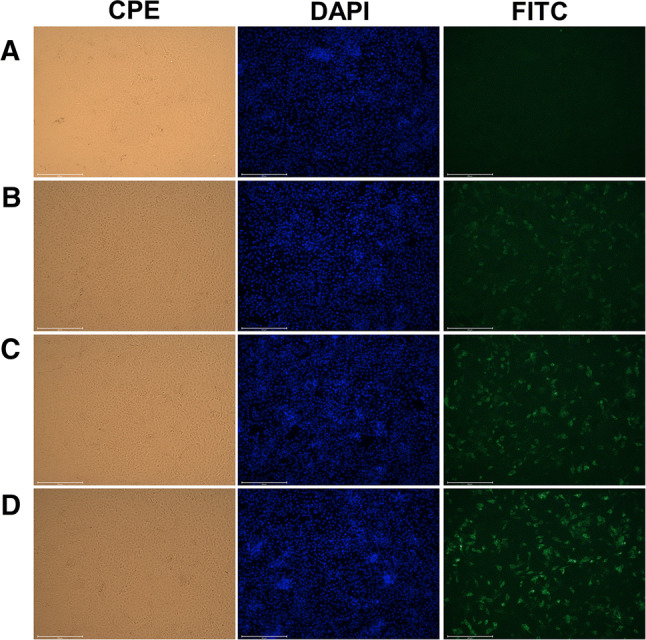
Identification of PDCoV-negative and positive sera by immunofluorescence assay. A. Negative serum samples showed no visible fluorescence in the immunofluorescence assay. B. Weakly positive serum samples showed low-intensity fluorescence. C. Strongly positive serum samples showed high-intensity fluorescence. D. Porcine anti-PDCoV hyperimmune serum was used as a positive control. (200× magnification)
Fig. 5.
ROC curve using the IFA as the diagnostic standard
Detection of PDCoV in field samples
A total of 152 serum and 62 milk samples were collected from six provinces and tested using the PDCoV S1 IgA ELISA. The results showed that 62.5% (95/152) of the serum samples and 100% (65/65) of the milk samples were positive for anti-PDCoV IgA antibodies (Table 1).
Table 1.
Rate of detection of anti-PDCoV IgA antibodies in pig serum and milk samples
| Province | Positive rate | Sample type |
|---|---|---|
| Heilongjiang | 57.5% (23/40) | Serum |
| Hebei | 42.5% (17/40) | Serum |
| Beijing | 75.0% (30/40) | Serum |
| Liaoning | 78.1% (25/32) | Serum |
| Hefei | 100% (46/46) | Milk |
| Tianjin | 100% (19/19) | Milk |
| Total | 73.73% (160/217) |
Discussion
PDCoV can cause vomiting and watery diarrhea in pigs, resulting in serious economic losses to the swine industry. Here, we have developed an indirect anti-PDCoV IgA ELISA that used the purified PDCoV S1 protein as a coating antigen for the measurement of serum and milk IgA levels in pigs, and used this assay to determine the prevalence of PDCoV on a few pig farms in China that had experienced diarrhea outbreaks within the previous two years.
In PDCoV-infected pigs, serum IgG and IgA antibodies could be detected at 14 days post-inoculation, after which PDCoV-specific IgG and IgA antibody titers increased and remained high until the pigs had fully recovered from clinical disease [28]. In our study, IgA-positive milk samples tested negative for IgG, indicating that the concentration of IgG antibodies in milk was relatively low or that the method was not sensitive enough to detect IgG antibodies. Based on what is known about IgA and IgG dynamics during PDCoV infection, we chose to detect IgA rather than IgG, which allows PDCoV infection to be monitored by detecting IgA antibodies in milk samples. PDCoV is an enteric pathogen that replicates primarily on mucosal surfaces. As with other coronaviruses, the presence of serum antibodies against PDCoV is not always correlated with protection. On the contrary, detection of antibodies only shows that individuals have had previous contact with pathogens or have been vaccinated [25–27].
In the present study, an indirect PDCoV IgA ELISA was developed to monitor IgA levels in serum and milk. This assay will aid in the future assessment of protection against PDCoV infection. Mucosal immunity plays an important role in immune protection, not only because it has a general pathogen-clearing effect but also because it involves both humoral and cellular immunity [29]. IgA is an important immunoglobulin for mucosal immunity that is secreted by the intestinal mucosa, and it provides resistance against invasive pathogens. High-affinity IgA can directly neutralize toxins and pathogens, while low-affinity IgA can prevent pathogen adhesion. IgA is also an important serum immunoglobulin that mediates a variety of protective functions through interactions with specific receptors and immune mediators [30].
PDCoV has an obvious intestinal tropism, and it is necessary to investigate the dynamics of PDCoV antibody production and antibody duration to better understand the mechanism of immunity to virus-induced diarrhea. Therefore, the indirect ELISA method established in this study provides a convenient tool for measuring serum and milk IgA levels in pigs as well as a basis for studying clinical IgA antibody responses to PDCoV infection and assessing the efficacy of future vaccines.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was funded by the National Key Research and Development Program of China (2016YFD0500705) and the Heilongjiang Province Natural Science Foundation of China (C2016065).
Compliance with ethical standards
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical approval
All procedures involving animals were done in accordance with ethical standards.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Manman Lu and Qiuge Liu contributed equally to this work.
Contributor Information
Jianfei Chen, Email: chenjianfei@caas.cn.
Li Feng, Email: fengli@caas.cn.
References
- 1.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen F, Zhu Y, Wu M, Ku X, Yao L, He Q. Full-length genome characterization of chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc. 2015;3(5):e01284-15. doi: 10.1128/genomeA.01284-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2(2):e00278-14. doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6(2):e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20(8):1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YW, Yue H, Fang W, Huang YW. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3(5):e00945-15. doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S, Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg Infect Dis. 2016;22(4):757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2(6):e01191-14. doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorsirigool A, Saeng-Chuto K, Temeeyasen G, Madapong A, Tripipat T, Wegner M, Tuntituvanont A, Intrakamhaeng M, Nilubol D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Adv Virol. 2016;161(10):2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, Zhang H, Lv DH, Li F, Wang D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Wang X, Jiao H, Shi H, Zhang X, Liu J, Shi D, Feng L. Isolation and identification of the first porcine deltacoronavirus strain in China. Chin J Prev Vet Med. 2016;38(3):171–174. [Google Scholar]
- 13.Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiou HY, Huang YL, Deng MC, Chang CY, Jeng CR, Tsai PS, Yang C, Pang VF, Chang HW. Phylogenetic analysis of the spike (S) gene of the new variants of porcine epidemic diarrhoea virus in Taiwan. Transbound Emerg Dis. 2017;64(1):157–166. doi: 10.1111/tbed.12357. [DOI] [PubMed] [Google Scholar]
- 15.Beniac DR, Andonov A, Grudeski E, Booth TF. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol. 2006;13(8):751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham S, Kienzle TE, Lapps W, Brian DA. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176(1):296–301. doi: 10.1016/0042-6822(90)90257-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64(11):5367–5375. doi: 10.1128/JVI.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luytjes W, Sturman LS, Bredenbeek PJ, Charite J, van der Zeijst BA, Horzinek MC, Spaan WJ. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161(2):479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuchel M, Ackermann M, Muller HK, Kihm U. An ELISA for detection of antibodies against porcine epidemic diarrhoea virus (PEDV) based on the specific solubility of the viral surface glycoprotein. Vet Microbiol. 1992;32(2):117–134. doi: 10.1016/0378-1135(92)90100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simkins RA, Weilnau PA, Van Cott J, Brim TA, Saif LJ. Competition ELISA, using monoclonal antibodies to the transmissible gastroenteritis virus (TGEV) S protein, for serologic differentiation of pigs infected with TGEV or porcine respiratory coronavirus. Am J Vet Res. 1993;54(2):254–259. [PubMed] [Google Scholar]
- 21.Elia G, Decaro N, Martella V, Lorusso E, Mari V, Maria SL, Cordioli P, Buonavoglia C. An ELISA based on recombinant spike protein S for the detection of antibodies to transmissible gastroenteritis virus of swine-like canine coronaviruses. J Virol Methods. 2010;163(2):309–312. doi: 10.1016/j.jviromet.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Zhang Y, Liang X, Oglesbee M, Krakowka S, Niehaus A, Wang G, Jia A, Song H, Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su M, Li C, Guo D, Wei S, Wang X, Geng Y, Yao S, Gao J, Wang E, Zhao X, Wang Z, Wang J, Wu R, Feng L, Sun D. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus. J Vet Med Sci. 2016;78(4):601–606. doi: 10.1292/jvms.15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thachil A, Gerber PF, Xiao CT, Huang YW, Opriessnig T. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies. PLoS One. 2015;10(4):e0124363. doi: 10.1371/journal.pone.0124363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saif LJ, van Cott JL, Brim TA. Immunity to transmissible gastroenteritis virus and porcine respiratory coronavirus infections in swine. Vet Immunol Immunopathol. 1994;43(1–3):89–97. doi: 10.1016/0165-2427(94)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saif LJ. Mucosal immunity: an overview and studies of enteric and respiratory coronavirus infections in a swine model of enteric disease. Vet Immunol Immunopathol. 1996;54(1–4):163–169. doi: 10.1016/S0165-2427(96)05702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.To TL, Ward LA, Yuan L, Saif LJ. Serum and intestinal isotype antibody responses and correlates of protective immunity to human rotavirus in a gnotobiotic pig model of disease. J Gen Virol. 1998;79(11):2661–2672. doi: 10.1099/0022-1317-79-11-2661. [DOI] [PubMed] [Google Scholar]
- 28.Hu H, Jung K, Vlasova AN, Saif LJ. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Adv Virol. 2016;161(12):3421–3434. doi: 10.1007/s00705-016-3056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Arriba ML, Carvajal A, Pozo J, Rubio P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet Immunol Immunopathol. 2002;85(1–2):85–97. doi: 10.1016/S0165-2427(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 30.Salmon H. Mammary gland immunology and neonate protection in pigs. Homing of lymphocytes into the MG. Adv Exp Med Biol. 2000;480:279–286. doi: 10.1007/0-306-46832-8_32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.