Abstract
In the present study, the detection of a pantropic canine coronavirus (CCoV) strain in a dog with lethal diarrhoea is reported. RT-PCR and real-time RT-PCR assays were used for the detection, characterization and quantitation of CCoV. Sequence and phylogenetic analysis of the CCoV NA/09 revealed a high degree of sequence identity with the pantropic strain CB/05, indicating the presence of CB/05-like pantropic strains in Greece. The absence of the 38-nucleotide deletion in ORF3b, which is characteristic of CB/05, indicates the need to identify new genetic markers for pantropic variants of CCoV, probably in the spike-protein gene region.
Keywords: CCoV, Pantropic, Dog, Genetic markers
Canine coronavirus (CCoV) is a member of the virus family Coronaviridae, order Nidovirales. It is a large, enveloped, single-stranded, positive-sense RNA virus. The genome consists of two overlapping open reading frames (ORF1a and 1b) translated into a polyprotein (precursor of the RNA-dependent RNA polymerase) and several ORFs encoding structural (spike, membrane, envelope and nucleocapsid protein) and non-structural proteins (3a, 3b, 3c, 7a and 7b) [14, 18]. To date, two different genotypes of CCoV have been recognized, CCoV type I (CCoV-I) and CCoV type II (CCoV-II) [10]. Furthermore, recently, CCoV-II was divided in two subtypes, CCoV-IIa and CCoV-IIb, with the second of these arising as a result of a putative recombination event between CCoV-II and transmissible gastroenteritis virus (TGEV) [12, 13]. Recently, CCoV, TGEV and feline coronavirus (FCoV) were included in the same species, Alphacoronavirus 1, of the new genus Alphacoronavirus [2].
CCoV is the causative agent of enteritis in dogs, with infection being restricted to the intestinal tract. Although CCoV is characterized by low mortality, and systemic infections are not usual [10], in 2005, a highly pathogenic CCoV strain (CB/05) was detected in an outbreak of a fatal systemic disease in puppies. The virus was isolated from faeces, as well as various parenchymatous organ samples from the dogs [1]. In addition, the disease was reproduced after experimental infection of dogs, with young puppies being more severely affected [11].
Sequencing and phylogenetic analysis have demonstrated that CB/05 belongs to the CCoV-IIa subtype, which includes all “classical” canine coronavirus strains, with a deletion in ORF3b being proposed as a possible genetic marker for the pantropic nature of the strain [10]. Recently, recombinant CCoV-IIb strains were also detected in organs of dogs, though after experimental infection, no systemic signs or virus dissemination through the blood were observed, implying differences in pathobiology among CCoV-IIa pantropic variants and CCoV-IIb strains [12, 23].
In Greece, the presence of CCoV was recently reported for the first time, in a severe outbreak of enteritis in a kennel. By sequence and phylogenetic analysis, high similarity to the pantropic CB/05 strain in the most variable region of the S gene was demonstrated [22]. In the present study, the molecular characterization and sequence analysis findings of a CCoV-IIa pantropic strain, as well as the need for new genetic markers, are reported.
In April 2009, a dead 10-week-old Maltese (13/09) male dog was submitted for laboratory investigation, coming from a pet shop in Thessaloniki, a city in northern Greece. The dog presented inappetence, depression, severe haemorrhagic diarrhoea, vomiting and hypothermia following fever, leading to death one day after the onset of the symptoms. The puppy had been vaccinated with a single dose of a vaccine against canine parvovirus and canine distemper virus three weeks before the onset of the symptoms.
During necropsy examination, linear haemorrhages of the intestinal wall, haemorrhagic enteritis and an ulcerated duodenum were observed. Multiple areas of emphysema and congestion were found at the lungs. The liver was enlarged, friable and yellow-brown in color with multifocal discolored spots. Congested vessels in the dura mater of the brain were also observed.
Faecal and parenchymatous organ samples were tested, using molecular diagnostic assays described previously, for common canine viral pathogens e.g., CCoV [3], canine parvovirus type 2 (CPV-2) [4, 6, 7], canine distemper virus (CDV) [15] and canine adenovirus type 1 and type 2 (CAV-1 and CAV-2) [17]. For CCoV typing, quantitation and subtyping in faecal and organ samples, two real-time RT-PCR assays [5] and two RT-PCR assays [13] were carried out.
The lung of the dead pup was homogenised (10% w/v) in D-MEM together with antibiotics (penicillin 1,000 IU/ml, streptomycin 100 μg/ml), and inoculated into canine fibroma (A-72) cells. Infected cells were monitored daily for the occurrence of cytopathic effect (CPE) and, after five days of incubation, they were tested for CCoV antigen by an immunofluorescence (IF) assay using a cat polyclonal serum specific for members of the species Alphacoronavirus 1 and a goat anti-cat IgG conjugated with fluorescein isothiocyanate (Sigma Aldrich, USA).
The 3′ end of the genome of the CCoV-IIa NA/09 strain was amplified as described previously, using viral RNA extracted from the spleen, SuperScript One-Step RT-PCR for Long Templates (Invitrogen s.r.l., Italy) and six RT-PCR assays, specific for overlapping fragments, encompassing ORFs 2, 3a, 3b, 3c, 4, 5, 6, 7a and 7b [9]. The nucleotide sequence was determined in both directions for each fragment. PCR amplicons were sequenced by a commercial facility (Beckman Coulter Genomics, United Kingdom). Sequence assembly and analysis were carried out using the BioEdit software package [16] and the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) analysis tool. The NetNGlyk 1.0 Server was used for the prediction of N-linked glycosylation sites, signal peptides and transmembrane helices (http://www.cbs.dtu.dk/services/, NetGlyc, SignalP and TMHMM, respectively). Phylogenetic analysis was conducted using the MEGA4 program, and a tree based on the S protein was constructed using the neighbor-joining method [24].
CCoV RNA was detected in the faeces, lungs, spleen, kidneys, pancreas, heart, liver and brain of the dead puppy. Using genotype-specific real-time RT-PCR assays, both genotypes, CCoV-I and CCoV-II (5.3 × 103 and 6.13 × 105 RNA copies/μl of template, respectively), were detected in the faeces, while only CCoV-II was detected in all organ samples. In detail, CCoV-II was detected in liver (5.11 × 103 RNA copies/μl of template), spleen (1.06 × 105 RNA copies/μl of template), pancreas (7.03 × 101 RNA copies/μl of template), kidney (1.21 × 103 RNA copies/μl of template), lungs (7.68 × 104 RNA copies/μl of template), heart (8.2 × 104 RNA copies/μl of template) and brain (2.03 × 102 RNA copies/μl of template). Regarding subtype characterization, the CCoV-II strain detected in faecal and in all organ samples was characterized as CCoV-IIa.
Despite several attempts, the virus was not adapted to growth in vitro, as demonstrated by the absence of cytopathic effect in inoculated cells and by negative results in the IF assay, even after three passages on cell cultures.
Regarding other canine viral pathogens, the dog tested positive for a CPV-2a field strain.
A total of 8,809 nucleotides were determined for strain NA/09, encompassing ORFs 2 (S protein), 3a, 3b, 3c, 4 (E protein), 5 (M protein), 6 (N protein), 7a and 7b. Alignment of an 8,767-nucleotide part of the sequence with TGEV, CCoV and FCoV reference strains available in GenBank showed the highest identity to CCoV-IIa pantropic reference strain CB/05 (DQ112226) (97.7%). The minimum conserved signal for transcription (CTAAAC) was present upstream of all of the ORFs (ORF 2, 3, 4, 5, 6 and 7).
The spike protein gene was 4,365 nucleotides long, encoding a protein of 1,454 amino acids. When compared to strain CB/05, no insertions or deletions were observed. The spike protein of NA/09, when compared with the analogous proteins of CCoV, TGEV and FCoV reference strains, showed the highest similarity to CB/05 (98.2% aa) (Table 1). Analysis using the NetNGlyk 1.0 Server showed that the protein contained a putative signal peptide (cleavage sites at positions Ser 19 to Thr 20, as in strain CB/05) and a transmembrane domain (positions 1,394 - 1,416 of the S gene). The S protein contained 31 potential N-linked glycosylation sites. Phylogenetic analysis revealed that the Greek strain was more closely related to the CB/05 pantropic reference strain detected in dogs’ organs (Fig. 1).
Table 1.
NA/09 S protein similarities, compared to S proteins of CCoV, TGEV and FCoV reference strains
| Virus strain | % Similarity to the NA/09 S protein |
|---|---|
| CCoV-IIa/CB/05/DQ112226 | 98.2 |
| CCoV-IIa/1-71/AY796289 | 94 |
| CCoV-IIa/K378/X77047 | 93.3 |
| CCoV-IIa/INSAVC/D13096 | 92.5 |
| CCoV-IIa/BGF10/AY342160 | 92 |
| CCoV-I/23/03/AY307021 | 43.6 |
| CCoV-I/Elmo/02/AY307020 | 42.9 |
| CCoV-IIb/341/05/EU856361 | 83.3 |
| CCoV-IIb/119/08/EU924791 | 83.7 |
| CCoV-IIb/430/07/EU924790 | 83.2 |
| CCoV-IIb/174/06/EU856362 | 82 |
| FCoV-I/UCD1/AB088222 | 43.2 |
| FCoV-I/Black/EU186072 | 42.6 |
| FCoV-II/79-1683/X80799 | 96 |
| FCoV-II/79-1146/NC007025 | 94.4 |
| TGEV/TS/DQ201447 | 80.3 |
| TGEV/Purdue/NC002306 | 80.3 |
Fig. 1.
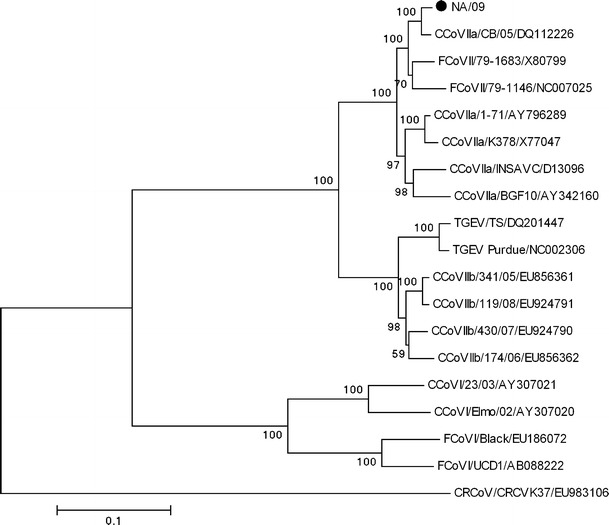
Neighbor-joining tree of the Greek strain, based on the S protein. The tree is rooted on the group 2 canine respiratory coronavirus (CRCoV). The numbers represent the percentage of replicate trees based on 1,000 bootstrap replicates. The scale bar represents the number of amino acid substitutions
The genomic region located between the S and E genes of both strains was found to encode three presumptive non-structural proteins. Nsp3a, 3b and 3c proteins were 78, 71 and 244 amino acids long, respectively. Regarding ORF3b, the 38-nucleotide deletion of the CB/05 was not observed (Fig. 2).
Fig. 2.

ORF3b nucleotide sequence of the NA/09 strain, aligned with CCoV-IIa reference strains CB/05, INSAVC, BGF10 and 1-71. The nucleotide deletions of CB/05 and 1-71 are enclosed in a rectangle
The envelope protein was found to be 82 amino acids in length, like in most canine coronavirus strains. A transmembrane domain was predicted between residue 15 and 37. The Greek strain had 100% amino acid identity to CB/05 and to two CCoV-IIb strains detected in dogs’ organs (119/08 and 341/05). The membrane protein (M protein) was found to be 262 amino acids long. It contained three potential N-linked glycosylation sites, a putative signal peptide (cleavage sites at positions Gly 16 to Glu 17) and three transmembrane domains (positions 47-69, 76-98 and 113-135). The M protein had the highest amino acid sequence identity to those of the CCoV-IIb reference strains (119/08 and 430/07) and to that of CB/05 (99.6% and 99.2%, respectively). The N protein (nucleoprotein) was found to be 382 amino acids in length. The Greek strain shared the highest amino acid identity with CCoV-IIb 119/08 and CB/05 (99.7% and 99.4%, respectively). ORF7 of both strains was found to encode two presumptive non-structural proteins. Nsp7a and 7b were 101 and 213 amino acids in length, and they were conserved with respect to the CCoV-II reference strains.
Coronaviruses are characterized by high genetic diversity. The high frequency of mutations, due to the high rate of mistakes of the RNA-dependent RNA polymerase, as well as frequent recombination are considered the main mechanisms of coronaviruses evolution, leading to the emergence of new strains, subtypes and genotypes within species and changes regarding tissue or even host tropism [26]. In the present study, a CCoV strain, spreading to the internal organs, was detected and analysed by molecular techniques. By means of sequence and phylogenetic analysis, a high similarity to the pantropic CB/05 variant was shown. This study, the second one reporting the genetic characterization of a CCoV pantropic strain, suggests that pantropic CCoV variants are present in Greece and can be associated with fatal disease in puppies [9]. However, the observed mortality could be due to the dual CPV/pantropic CCoV infection, considering that mixed infections usually display a more severe course and higher mortality [8]. In addition, recent studies have demonstrated that the outcome of CB/05 infection is not invariably lethal, but, at least under experimental conditions, its main consequence is represented by a profound, long-lasting lymphopenia involving both B and T cells [20].
By means of real-time RT-PCR, the tissue distribution of NA/09 was determined. Viral RNA was detected in several parenchymatous organs and in high concentration, suggesting the pantropic nature of the variant. Detection and quantitation of the viral RNA in the heart and the pancreas is reported for the first time. The detection of CCoV-I, which was restricted to the faecal samples, suggests that the pantropic NA/09 strain has the advantage of disseminating in the organism, revealing differences in the pathobiology of the pantropic variants with respect to common CCoV enteric strains.
Regarding CCoV, mutations are considered to play a key role in the emergence of the pantropic variant CB/05 [9]. The truncated form of ORF3b, observed in CB/05, was not present in NA/09. That mutation was considered to be a possible genetic marker for pantropic variants [9]. However, similar deletions in ORF3b have been observed in common enteric strains, as in 1-71 (Fig. 2) [19], and truncated nsp3b has been associated with attenuated coronavirus strains [25]. On the other hand, a high similarity in the S protein was observed between NA/09 and CB/05 strains. Since the S protein is responsible for receptor attachment and changes in tissue tropism have been associated with mutations in the S gene [18, 21], it is likely that specific genetic markers for pantropic strains will be identified in this region.
In conclusion, in the present study, the detection, quantitation and molecular analysis of a pantropic CCoV variant is reported. In addition, it is suggested that pantropic variants can be identified in the heart and the pancreas. The emergence of strains that are able to disseminate and cause systemic infection suggests that canine coronaviral infections may be more interesting and severe than thought in the past. Further studies are necessary in order to determine the circulation of pantropic CCoV strains among the dog population and to identify specific genetic markers that can be useful for understanding virus pathobiology and epidemiology.
Acknowledgments
Ntafis Vasileios is grateful to Alexander S. Onassis Public Benefit Foundation for doctoral funding.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Nucleotide sequence data reported is available in the GenBank database under the accession number JF682842.
References
- 1.Buonavoglia C, Decaro N, Martella V, Elia G, Campolo M, Desario C, Castagnaro M, Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decaro N, Pratelli A, Campolo M, Elia G, Martella V, Tempesta M, Buonavoglia C. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J Virol Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decaro N, Elia G, Martella V, Desario C, Campolo M, Di Trani L, Tarsitano E, Tempesta M, Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Decaro N, Martella V, Ricci D, Elia G, Desario C, Campolo M, Cavaliere N, Trani LD, Tempesta M, Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and the quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. J Virol Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decaro N, Elia G, Desario C, Roperto S, Martella V, Campolo M, Lorusso A, Cavalli A, Buonavoglia C. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J Virol Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decaro N, Elia G, Martella V, Campolo M, Desario C, Camero M, Cirone F, Lorusso E, Lucente MS, Narcisi D, Scalia P, Buonavoglia C. Characterization of the canine parvovirus type 2 variants using minor groove binder probe technology. J Virol Methods. 2006;133:92–99. doi: 10.1016/j.jviromet.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Decaro N, Martella V, Desario C, Bellacicco AL, Camero M, Manna L, D’aloja D, Buonavoglia C. First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. J Vet Med B Infect Dis Vet Public Health. 2006;53:468–472. doi: 10.1111/j.1439-0450.2006.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decaro N, Martella V, Elia G, Campolo M, Desario C, Cirone F, Tempesta M, Buonavoglia C. Molecular characterization of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:54–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decaro N, Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decaro N, Campolo M, Lorusso A, Desario C, Mari V, Colaianni ML, Elia G, Martella V, Buonavoglia C. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Vet Microbiol. 2008;128:253–260. doi: 10.1016/j.vetmic.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, Martella V, Cordioli P, Enjuanes L, Buonavoglia C. Recombinant canine coronavirus related to transmissible gastroenteritis virus of swine are circulating in dogs. J Virol. 2009;83(3):1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decaro N, Mari V, Elia G, Addie DD, Camero M, Lucente MS, Martella V, Buonavoglia C. Recombinant canine coronaviruses in dogs, Europe. Emerg Infect Dis. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enjuanes L, Brian D, Cavanagh D, Holmes K, Lai MMC, Laude H, Masters P, Rottier PJM, Siddell SG, Spaan WJM, Taguchi F, Talbot P, et al. Coronaviridae. In: van Regenmortel MHV, et al., editors. Virus taxonomy. Classification and nomenclature of viruses. New York: Academic Press; 2000. pp. 835–849. [Google Scholar]
- 15.Frisk AL, Konig M, Moritz A, Baumgartner W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J Clin Microbiol. 1999;37:3634–3643. doi: 10.1128/jcm.37.11.3634-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp. 1999;41:95–98. [Google Scholar]
- 17.Hu RL, Huang G, Qiu W, Zhong ZH, Xia XZ, Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet Res Commun. 2001;25:77–84. doi: 10.1023/A:1006417203856. [DOI] [PubMed] [Google Scholar]
- 18.Lai MMC, Holmes KV, et al. Coronaviridae: the viruses and their replication. In: Knipe DM, et al., editors. Fields virology. 4. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 1163–1185. [Google Scholar]
- 19.Ma G, Wang Y, Lu C. Molecular characterization of the 9.36kb C-terminal region of canine coronavirus 1–71 strain. Virus genes. 2008;36:491–497. doi: 10.1007/s11262-008-0214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marinaro M, Mari V, Bellacicco AL, Tarsitano E, Elia G, Losurdo M, Rezza G, Buonavoglia C, Decaro N. Long-lasting depletion of circulating CD4+ T lymphocytes and acute monocytosis after pantropic canine coronavirus infection in dogs. Virus Res. 2010;152:73–78. doi: 10.1016/j.virusres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ntafis V, Mari V, Danika S, Fragkiadaki E, Buonavoglia C. An outbreak of canine coronavirus in a Greek kennel. J Vet Diagn Invest. 2010;22:320–323. doi: 10.1177/104063871002200231. [DOI] [PubMed] [Google Scholar]
- 23.Ntafis V, Mari V, Decaro N, Papanastassopoulou M, Papaioannou N, Mpatziou R, Buonavoglia C, Xylouri E. Isolation, tissue distribution and molecular characterization of two recombinant canine coronavirus strains. Vet Microbiol. 2011;151:238–244. doi: 10.1016/j.vetmic.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Wesley RD, Woods RD, Cheung AK. Genetic basis for the pathogenicity of transmissible gastroenteritis virus. J Virol. 1990;64:4761–4766. doi: 10.1128/jvi.64.10.4761-4766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo PCY, Lau SKP, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]


