Abstract
Four types of porcine epidemic diarrhea virus (PEDV) variants with a large deletion in the spike protein were detected, together with the original US PEDV, from pig fecal and oral fluid samples collected during 2016-2017 in the US. Two of the variants are similar to those identified in Japan: one contains a 194-aa deletion, the same as PEDV variant TTR-2/JPN/2014, while the other contains a 204-aa deletion, the same as PEDV variant JKa-292/CS1de204. Two new S1 NTD-del PEDV variants were found: one contains a 201-aa deletion located at residues 30-230 and the other contains a 202-aa deletion located at residues 24-225 of the S protein. This is the first report on coinfection of S1 NTD-del PEDV variants and the original US PEDV strain in US pigs, indicating that PEDV continues to evolve in pigs and might be responsible for disease pattern changes.
In spring 2013, porcine epidemic diarrhea virus (PEDV), an alphacoronavirus, emerged for the first time in the US, causing large-scale outbreaks in swine farms and the deaths of seven million pigs in the first year as well as severe economic losses [4]. Since summer 2014, the porcine epidemic diarrhea (PED) disease pattern has been changing; while epidemics have decreased, sporadic outbreaks still occur and the virus is becoming more persistent at some farms. Two major types of PEDV, the original US [or non-S INDEL or North American (NA)] and S INDEL (G1b), are co-circulating in the US. By summer 2017, PEDV had been reported in 39 states.
We detected the first S INDEL strain of PEDV in the US and isolated several original US and S INDEL PEDV strains and a unique variant in Vero cell cultures [6, 11]. The unique strain is the tissue culture-adapted (TC) PC177 strain that bears a 197-amino-acid (aa) deletion in the N-terminal domain (NTD) of the spike (S) protein. We designated it as a S1 NTD-del variant [2]. We did not detect any PEDV viral RNA harboring this deletion in the original field samples (or wild-type) of PC177 and concluded that the large deletion evolved during Vero cell culture [6]. Later, we developed new RT-PCR assays to differentiate the original US, S INDEL and S1 NTD-del PEDV variants [5]. We re-tested the original PC177 sample and screened dozens of PEDV-positive fecal samples previously tested positive by TaqMan real-time RT-PCR (RT-qPCR) targeting the nucleocapsid (N) gene of PEDV [6]. Those samples were collected from PED outbreaks in Ohio during May 2013 and February 2014. We still did not detect an S1 NTD-del PEDV. In this study, we screened clinical pig samples from April 2014 to May 2017 and detected the S1 NTD-del PEDV variants in the US as early as December, 2016.
Twenty-nine PEDV-positive pig fecal/intestine and oral fluid samples were provided by the Animal Disease Diagnostic Laboratory (ADDL) of the Ohio Department of Agriculture (Table 1). RNA extraction, cDNA synthesis and PCR assays with primer sets F1 (5’-ATGGTACGTTGCTAGTGCGTA-3’) and UR (5’-TACCATGCACCATAGTGGAATCAT-3’), F2 (5’-TGGTGAAATCCAGAGTGTCAAT-3’) and UR, and F3 (5’-CCAGCTTATATGCAGGATGGAA-3’) and UR were performed to differentiate S1 NTD-del, original US and S INDEL PEDV as described previously (Table 1) [5]. TC-PC177, the original US PEDV TC-PC22A, and S INDEL PEDV TC-Iowa106 strains were used as positive controls [6]. Distilled water was used as a negative control.
Table 1.
Detection of S1 NTD-del PEDVs from PEDV-positive pig fecal/oral fluid samples
| Sample # | Collected time (M/D/Y) | Nature | Age of pig(s) | Clinical Signs | PCR w/F1/UR (bp)a | PCR w/F2/UR (bp)a | PCR w/F3/UR (bp)a | Sequence analysis | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|
| PC245 | 4/23/2014 | Feces | <1 month | Yesf | ~ 880 | ~ 650 | - | NDe | |
| PC246 | 5/14/2014 | Feces | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC247 | 1/8/2015 | Intestine | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC248 | 1/23/2015 | Intestine | <1 month | Yesf | ~ 880 | ~ 650 | ~358 | ND | |
| PC249 | 1/23/2015 | Intestine | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC250 | 5/8/2015 | Rectal swab | 1-3 months | Yesf | ~ 880 | - | - | Original USd | |
| PC251 | 11/2/2015 | Intestine | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC252 | 5/13/2016 | Oral fluid | 1-3 months | No | ~ 880 | ~ 650 | - | ND | |
| PC253 | 5/13/2016 | Feces | 1-3 months | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC254 | 4/27/2016 | Intestine | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC255 | 4/27/2016 | Rectal swab | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC256 | 12/2/2016 | Feces | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC257 | 12/2/2016 | Feces | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC258 | 12/6/2016 | Oral Fluid | Unknownh | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC259/O | 12/8/2016 | Oral Fluid | Unknownh | Yesf | ~ 880; | ND | ND | Original US; | |
| PC259/D194 | Unknownh | ~ 302 | S1 NTD-Del | MG490982 | |||||
| PC260 | 12/29/2016 | Feces | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC261/Ob | 12/29/2016 | Feces | 3-8 months | Yesf | ~ 880; | ND | ND | Original US; | |
| PC261/D194b | ~ 302 | S1 NTD-Del | MG837056 | ||||||
| PC261/D201b | MG837057 | ||||||||
| PC262/Ob | 1/19/2017 | Fecal Swab | 1-3 months | Yesf | ~ 880; | ND | ND | Original US; | MG490984 |
| PC262/D194b | ~ 302 | S1 NTD-Del | MG490983 | ||||||
| PC263 | 12/30/2016 | Oral fluid | 3-8 months | Mildg | ~ 880 | ~ 650 | - | ND | |
| PC264 | 1/20/2017 | Rectal swab | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC265 | 1/20/2017 | Rectal swab | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC266 | 1/27/2017 | Oral fluid | Unknownh | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC267 | 2/10/2017 | Fecal swab | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC268 | 2/23/2017 | Fecal swab | <1 month | Yesf | ~ 880 | ~ 650 | - | ND | |
| PC269 | 2/31/17 | Feces | 3-8 months | Mildg | ~ 880 | - | - | Original US d | |
| PC270 | 3/17/2017 | Oral fluid | <1 month | Yesf | ~ 880 | - | ~ 358 | S INDEL | |
| PC271 | 3/31/2017 | Oral fluid | 1-3 months | Yesf | ~ 880 | - | - | Original US d | |
| PC272/Oc | 5/13/2017 | Oral fluid | <1 month | Yesf | ~ 880; | ND | ND | Original US; | |
| PC272/D194c | ~ 302 | S1 NTD-Del | MG490985 | ||||||
| PC273/Oc | 5/31/2017 | Oral fluid | <1 month | Yesf | ~ 880; | ND | ND | Original US; | MG837058 |
| PC273/D194c | ~ 302 | S1 NTD-Del | MH000205 | ||||||
| PC273/D202c | MH000206 | ||||||||
| PC273/D204c | MH000207 |
aPrimer set F1/UR is for the detection of all three types of PEDV with different amplicon sizes [original US and S INDEL PEDV (~880 bp); S1 NTD-del PEDV (~302 bp)]; F2/UR is for the original US PEDV (~650 bp); and F3/UR is for S INDEL PEDV (~358 bp)
bThese two samples (PC261 and PC262) were collected from the same farm at different times
cThese two samples (PC272 and PC273) were collected from the same farm at different times
dOriginal US PEDV strains with nucleotide mutations at the binding site or near the 3’-end of primer F2
eND: not done
fAcute watery diarrhea with no blood or mucus, increased mortality
g“showing signs of looseness”
h‘Unknown’: Submitting veterinarian did not include age of pigs
-: Negative
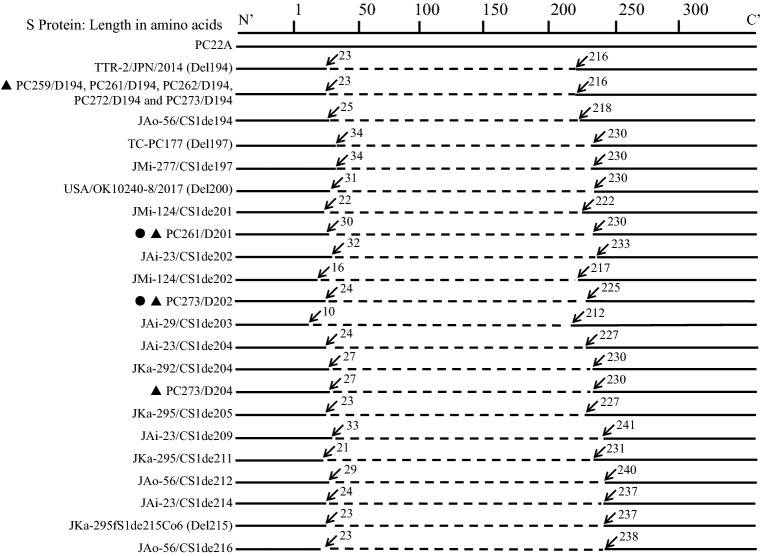
Among the 29 samples, 24 showed a single 880 bp-band with primer set F1/UR, indicating that they were positive for the original US PEDV or S INDEL PEDV, while five samples (PC259, PC261, PC262, PC272 and PC273) showed both the 880 bp-band and the 302 bp-band, indicating that they were coinfected with original US PEDV/S INDEL PEDV and S1 NTD-del PEDV (Fig. 1a). Both bands of the five samples were purified and sequenced using Sanger sequencing method. After running Basic Local Alignment Search Tool (BLAST) at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), we found that the virus-specific sequences shared the highest nucleotide identities (97-99%) with the original US PEDV and S1 NTD-del PEDV, respectively. Here, we designated the original US PEDV in a sample as ‘sample no./O’ and S1 NTD-del PEDV as ‘sample no./D followed by the number of amino acid deletions’. The deletion region of samples PC259/D194, PC262/D194 and PC272/D194 is the same, which is a 194 aa-fragment located between residues 23-216 of the S protein (Fig. 2). It is the same as that of the first field S1 NTD-del PEDV variant TTR-2/JPN/2014 (LC063828), which was detected from a PED outbreak in October 2014 in Japan [8]. However, by cloning and sequencing the shorter PCR products (~ 900 bp) of samples PC261 and PC273 with the primer set PEDV-20320-F (5’-AACACGTCATCGTCAGAGGC-3’) and PEDV-21816R (5’-CGGTTGGAGGTAAAACAGC-3’) targeting the 3’-end of the nsp16 gene and S1 gene, respectively [6], we found that sample PC261 contained two types of S1 NTD-del variants: PC261/D194 contained the above 194-aa deletion, while PC261/D201 contained a novel 201-aa deletion located at residues 30-230 of the S protein (Fig. 2). Sample PC273 contained three types of S1 NTD-del variants: 1) PC273/D194 contained the above 194-aa deletion, 2) PC273/D204 contained a 204-aa deletion located between residues 27-230 of the S protein, which is as same as PEDV variant JKa-292/CS1de204 (KU363094) reported in Japan in 2017 [1], and 3) PC273/D202 contained a novel 202-aa deletion located at residues 24-225 of the S protein (Fig. 2). Additionally, we sequenced the complete genome of one sample (PC273/O) using next generation sequencing as described previously [11]. From our study, the S1 NTD-del PEDV variants appeared as early as December 2016 and continued to circulate to at least May 2017 in Ohio swine farms. The 24 samples with a single 880 bp-band were tested by PCR with primer sets F2/UR for the original US PEDV and F3/UR for S INDEL PEDV, respectively (Fig. 1b).
Fig. 1.
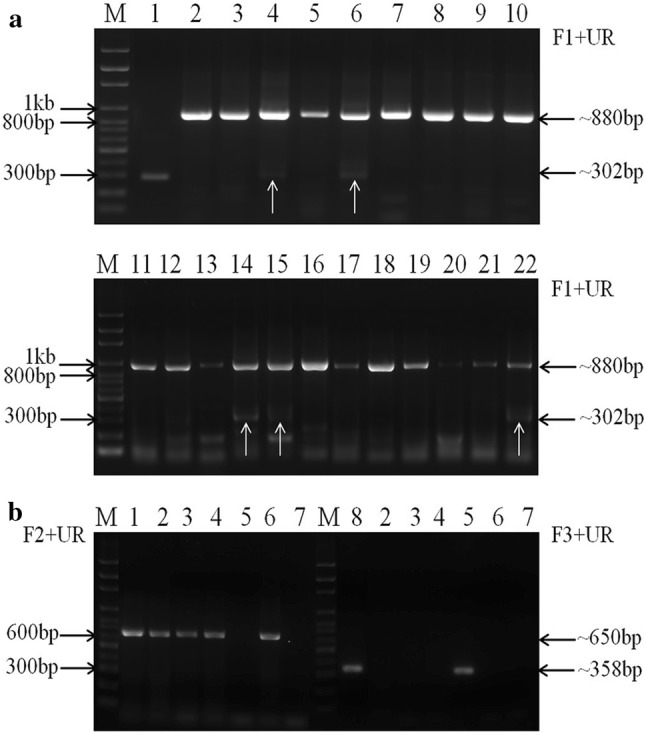
Differentiation of original US PEDV, S INDEL PEDV, and S1 NTD-del PEDV using PCR assays with primer sets F1/UR, F2/UR and F3/UR. (a) PCR products following primary amplification with primer set F1/UR for S1 NTD-del PEDV (~302 bp), original US PEDV (~880 bp) and S INDEL PEDV (~880 bp). Lane 1: TC-PC177 as a positive control for S1 NTD-del PEDV. Lane 2: TC-PC22A as a positive control for the original US PEDV. Lanes 3- 22: PCR products of some samples with primer set F1/UR. The white arrows refer to the positive bands for S1 NTD-del PEDV variants: Samples PC259, PC261, PC262, PC272 and PC273 correspond to lanes 14, 15, 4, 22 and 6, respectively. (b) Samples, which were amplified with a single 880 bp-band by PCR with primer set F1/UR, were tested with primer sets F2/UR and F3/UR for differentiation of the original US (~650bp) and S INDEL (~358bp) PEDV, respectively. Lane 1: TC-PC22A as a positive control for the original US PEDV. Lane 8: TC-Iowa106 as a positive control for S INDEL PEDV. Lanes 2-7: PCR products of some samples. M: 100 bp ladder DNA marker (PHENIX Research Products, NC, USA)
Fig. 2.
Diagram of deletion size and the location of each type of S1 NTD-del PEDV variant. PEDV variants detected in this study are marked with a solid triangle. Solid circles indicate the two new S1 NTD-del PEDV variants identified in this study
Among the 24 samples, 19 samples were positive for only the original US PEDV (650 bp). One sample (PC270) was positive only for S INDEL PEDV (358 bp). One sample (PC248) was positive for both types of PEDV. Three samples (PC250, PC269, and PC271) were positive with primer set F1/UR, but negative with both primer sets F2/UR and F3/UR. Therefore, their F1/UR PCR products (~880 bp) were sequenced. They were the original US PEDV with nucleotide mutations at the binding site or near the 3’-end of primer F2, resulting in no amplification by primer set F2/UR. In addition, we tried but failed to isolate any PEDV variants from the PEDV-positive clinical samples in Vero cells (ATCC#CCL81) using materials and methods described by our lab previously [6].
The S protein of a coronavirus is divided into S1 and S2 subunits, responsible for receptor binding and virus-cell membrane fusion to initiate infection, respectively [3]. The S1 subunit of an original US PEDV forms five domains (domain 0 [corresponding to the NTD], A, B, C and D) based on structure analysis [2]. Domain 0 and domain A are structurally similar and it has been hypothesized that either of them is likely dispensable [10]. It seems correct for PEDV because TC-PC177, TC-TTR-2, and an infectious clone-derived S1 NTD-del PEDV (icPC22A-S1Δ197) can replicate in vivo and in vitro [2, 9]. During the revision of this manuscript, a new publication reported that a single S1 NTD-del PEDV variant with a 200-aa del was detected from a diarrheic pig fecal sample in the US [12], indicating that PEDV with the S1 NTD-del PEDV is capable of replicating in live pigs. Why did the eight S1 NTD-del PEDV variants detected in this study co-infect pigs with the original US PEDV strains without exception, similar to the data from Japan? Fifteen types of S1 NTD-del PEDV variants bearing large deletions (ranging from 194 to 216 aa) (Fig. 2) co-infected pigs with PEDV strains possessing an intact S gene [1]. One possible reason could be that S1 NTD-del PEDV alone does not cause severe diarrhea as observed in the experimentally infected pigs [2, 9], and such samples were probably not submitted for diagnosis. The S1 subunit of the PEDV S gene is known to be the most variable region and has been widely used for studies of PEDV evolution and genetic diversity. By analyzing the partial sequences of S1 NTD-del PEDV variants in this study, we found the following facts: (1) The 258 nt sequences (covering from the 3’-end of the nsp16 gene to the partial S1 subunit) of PC262/O and PC262/D194 from the same clinical fecal sample had five single nucleotide polymorphisms (SNPs); (2) The 258 nt sequences (covering from the 3’-end of the nsp16 gene to the partial S1 subunit) of different clinical samples (PC259/D194, PC261/D194, PC262/D194, PC272/D194 and PC273/D194) had 0-7 SNPs, and the 858 nt sequences (covering from the 3’-end of the nsp16 gene to the partial S1 subunit) of PC261/D194 and PC273/D194 had six SNPs. Detection of both ‘D’ and ‘O’ variants in the same clinical sample indicates that the pig was infected with both viruses.
The NTD of S1 has been reported to be responsible for binding to sialic acids and the deletion of the S1 NTD did not decrease the induction of virus neutralizing antibodies, but reduced the virulence of the highly virulent PEDV in neonatal piglets [2]. It was documented that sialic acid binding activity of the PEDV S protein facilitates cell entry [3]. Therefore, although the S1 NTD of PEDV does not contain predominant virus neutralizing epitopes and seems dispensable, it is believed to be important for pathogenicity. A good example is the transmissible gastroenteritis virus (TGEV), another porcine enteric alphacoronavirus. Its S1 NTD-del variant porcine respiratory coronavirus (PRCV) changes the major tissue tropism of TGEV changing the disease from a primarily enteric one to a mild respiratory disease in pigs [7]. Due to the shared immunogenicity between TGEV and PRCV, the widespread prevalence of PRCV has lessened the clinical impact of TGE in Europe. Similarly, inoculation with S1 NTD-del PEDV is associated with decreased severity of enteric disease in pigs [2]. In contrast to TGEV, the S1 NTD-del PEDV strains retained their enteric tropism [2, 9]. Although the large scale outbreaks of PEDV in the US are now less commonly seen, sporadic small scale outbreaks or persistent infections of PEDV in some pig farms are still detected. We hypothesize that co-infection of S1 NTD-del PEDV variants may stimulate population immunity that protects the pigs from large scale outbreaks caused by the virulent strain of US PEDV. The variant strains may also be responsible for the persistence of virulent strains of PEDV at some farms. Further studies are warranted to determine if the clinically mild strains of S1 NTD-del PEDV variants continue to evolve and potentially replace the virulent strain of PEDV in the field, similar to what happened to PRCV and TGEV.
This is the first report of the coinfection of S1 NTD-del PEDV and original US PEDV strains in US pigs, indicating that active PEDV evolution is transpiring. This may contribute to the changing PED disease patterns in the US. The previously identified original US PEDV strains as well as the S INDEL PEDV are still circulating in the US.
Acknowledgements
We thank Xiaohong Wang, Patricia Boley, Katherine Shiplett, and Kerri Lawrence for technical assistance. Sanger sequencing were performed at Plant-Microbe Genomics Facility, and the Molecular and Cellular Imaging Center, the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University OSU. This work was partially funded by National Institute of Food and Agriculture, U.S. Department of Agriculture, under Award Number 2015-67015-23067 (QW, PI). Salaries and research support were provided by state and federal funds appropriated to OARDC, OSU. Yunfang Su’s stipend is supported by China Scholarship Council.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Contributor Information
Yan Zhang, Phone: 614-728-6220, Email: yan.zhang@agri.ohio.gov.
Qiuhong Wang, Phone: 330-263-3744, Email: wang.655@osu.edu.
References
- 1.Diep NV, Norimine J, Sueyoshi M, Lan NT, Yamaguchi R. Novel porcine epidemic diarrhea virus (PEDV) variants with large deletions in the spike (S) gene coexist with PEDV strains possessing an intact S gene in domestic pigs in Japan: a new disease situation. PloS one. 2017;12:e0170126. doi: 10.1371/journal.pone.0170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y, Lin CM, Yokoyama M, Yount BL, Marthaler D, Douglas AL, Ghimire S, Qin Y, Baric RS, Saif LJ, Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J Virol. 2017;91:e00227. doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch BJ. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Wang Q. Reverse transcription-PCR assays for the differentiation of various US porcine epidemic diarrhea virus strains. J Virol Methods. 2016;234:137–141. doi: 10.1016/j.jviromet.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oka T, Saif LJ, Marthaler D, Esseili MA, Meulia T, Lin CM, Vlasova AN, Jung K, Zhang Y, Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Veterinary Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saif LJ, Pensaert M, Sestak K, Yeo SG, Jung K. Coronaviruses. In: Zimmerman JJ, editor. Diseases of swine. Ames, IA: Wiley-Blackwell Publishing; 2012. pp. 501–524. [Google Scholar]
- 8.Suzuki T, Murakami S, Takahashi O, Kodera A, Masuda T, Itoh S, Miyazaki A, Ohashi S, Tsutsui T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect Genet Evolut. 2015;36:363–368. doi: 10.1016/j.meegid.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Shibahara T, Yamaguchi R, Nakade K, Yamamoto T, Miyazaki A, Ohashi S. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J Gen Virol. 2016;97:1823–1828. doi: 10.1099/jgv.0.000423. [DOI] [PubMed] [Google Scholar]
- 10.Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch BJ, Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Yim-Im W, Chen Q, Zheng Y, Schumacher L, Huang H, Gauger P, Harmon K, Li G. Identification of porcine epidemic diarrhea virus variant with a large spike gene deletion from a clinical swine sample in the United States. Virus Genes. 2018;54:323–327. doi: 10.1007/s11262-018-1542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]