Abstract
Porcine hemagglutinating encephalomyelitis virus (PHEV) is a typical neurotropic coronavirus that mainly invades the central nervous system (CNS) in piglets and causes vomiting and wasting disease. Emerging evidence suggests that PHEV alters microRNA (miRNA) expression profiles, and miRNA has also been postulated to be involved in its pathogenesis, but the mechanisms underlying this process have not been fully explored. In this study, we found that PHEV infection upregulates miR-142a-3p RNA expression in N2a cells and in the CNS of mice. Downregulation of miR-142a-3p by an miRNA inhibitor led to a significant repression of viral proliferation, implying that it acts as a positive regulator of PHEV proliferation. Using a dual-luciferase reporter assay, miR-142a-3p was found to bind directly bound to the 3’ untranslated region (3’UTR) of Rab3a mRNA and downregulate its expression. Knockdown of Rab3a expression by transfection with an miR-142a-3p mimic or Rab3a siRNA significantly increased PHEV replication in N2a cells. Conversely, the use of an miR-142a-3p inhibitor or overexpression of Rab3a resulted in a marked restriction of viral production at both the mRNA and protein level. Our data demonstrate that miR-142a-3p promotes PHEV proliferation by directly targeting Rab3a mRNA, and this provides new insights into the mechanisms of PHEV-related pathogenesis and virus-host interactions.
Introduction
Porcine hemagglutinating encephalomyelitis virus (PHEV) is a member of the genus Coronavirus, family Coronaviridae, that mainly causes vomiting and wasting disease (VWD) and/or encephalomyelitis in piglets under 3 weeks old [1, 2]. The disease was first reported in 1958, and the virus was first isolated from the piglets in 1962 [3, 4]. Since then, PHEV infection has been widespread worldwide, including Europe, Asia, and North America, where the overall mortality rate in piglets ranges from 30% to 100% [5–8]. Recently, an acute outbreak of influenza-like illness (ILI) in adult pigs in the USA was reported, and testing identified PHEV as the cause of the disease [9]. The ILI that was observed may reflect an atypical form of illness or a variant of PHEV with increased virulence. PHEV is a neurovirulent virus that spreads to the central nervous system (CNS) via peripheral nerves, where neurite damage and neuronal death are observed, causing neurological symptoms. Since pigs are the natural hosts of PHEV, the virus has to adapt to replicate in rats and mice (i.e., ICR, BALB/c and Wistar) in vivo, and rat dorsal root ganglia (DRG) and mouse neuroblastoma N2a cells in vitro [8, 10, 11]. The mechanism by which neuronal degeneration is induced by PHEV infection is not well understood, and the study of virus-host interactions is therefore important.
MicroRNAs (miRNAs) are single-stranded, conserved, endogenous non-coding RNAs (ncRNAs) that are 19-25 nucleotides (nt) in length and can regulate target gene mRNA expression through translational repression or degradation [12]. Several studies have shown that aberrant expression of miRNAs is closely associated with proliferation, invasion, metastasis and the prognosis of various coronavirus infections [13–15]. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection upregulates miR-17*, mir-574-5p, and miR-214 and inhibits the expression of miR-98 and miR-223. miR-17* and mir-574-5p have been shown to restrict the replication of SARS-CoV, whereas miR-214 contributes to immune escape [15]. miRNAs are processed from hairpin transcripts, and this maturation step gives rise to a 3p and a 5p strand, both of which allow the silencing of specific target genes through base pairing to a minimal recognition sequence [16]. The miR-142(a) hairpin gives rise to a “guide strand”, miR-142(a)-3p, and a sister “passenger strand”, miR-142(a)-5p [12]. miR-142a-3p has been sown to play a critical role in virus infection, inflammation, and immune tolerance. Trobaugh et al. showed that North American eastern equine encephalitis virus (EEEV) hijacks host miR-142-3p to limit viral replication in particular cell lines [17]. miR-142a-3p also modulates neutrophil development by controlling its maturation in zebrafish [18]. Using miRNA array, miR-142a-3p expression has been shown to be upregulated during PHEV infection [19]. However, it remains to be investigated whether miR-142a-3p controls virus proliferation or translation of mRNAs that are associated with neurological dysfunction.
In this study, we examine the expression of miR-142a-3p in N2a cells and explored the relationship between PHEV and miR-142a-3p. To gain insight into the potential roles of miR-142a-3p, we identified one of its target genes and investigated its functional significance during PHEV infection. The data show that miR-142a-3p may be upregulated by PHEV and promote viral replication by directly targeting Rab3a. The results enhance our understanding of how these neurotropic viruses are involved in CNS dysfunction at the posttranscriptional level, providing a theoretical basis for a new therapeutic strategy for PHEV infection.
Materials and methods
Cells, virus, and animals
Mouse neuroblastoma N2a (N2a) cells and human embryonic kidney (HEK293T) cells were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. The PHEV stain (HEV 67N) (GenBank no. AY048917) was grown in N2a cells. BALB/c mice (3 weeks old) were provided by the Laboratory Animal Center of Jilin University. All animal studies were conducted according to experimental practices and standards approved by the Animal Welfare and Research Ethics Committee of the College of Veterinary Medicine, Jilin University, China (permission number KT201904002), following the recommendations of the Council for International Organization of Medical Sciences on Animal Experimentation.
Transfection and PHEV infection
The miR-142a-3p mimic, inhibitor, and NC (negative control) were purchased from RiboBio (Guangzhou, China). Their sequences were as follows: miR-142a-3p mimic, 5’- UGU AGU GUU UCC UAC UUU AUG GA -3’ (forward) and 3’- ACA UCA CAA AGG AUG AAA UAC CU -5’ (reverse); miR-142a-3p inhibitor, 5’- mUmCmC mAmUmA mAmAmG mUmAmG mGmAmA mAmCmA mCmUmA mCmA -3’; and NC, 5’- UUU GUA CUA CAC AAA AGU ACU G -3’ (forward) and 3’- AAA CAU GAU GUG UUU UCA UGA C -5’ (reverse). HEK293T or N2a cells were transfected with the miRNA mimic or NC using X-tremeGENE HP DNA Transfection Reagent (Roche, Sweden). Transfection with miRNA inhibitor was done electroporation as described by Khanna et al. [20].
For PHEV infection experiments, N2a cells were infected with PHEV (multiplicity of infection [MOI] = 10) at 12 hours post-transfection. Cells or supernatants from cell cultures were collected at 24, 36, and 48 hours postinfection (hpi). Twenty 3-week-old male BALB/c mice were randomly divided into two groups. Mice in the PHEV-infected group were inoculated with 100 μL PHEV (104.45 median tissue culture infectious doses [TCID50]/0.1 mL) by the intranasal route, and those in the control group were inoculated with PBS in the same manner. Mouse brain tissues were harvested at 3 and 5 days dpi, and expression levels of miR-142a-3p and PHEV were determined by quantitative real-time PCR (qRT-PCR) and Western blot assays.
qRT-PCR assay
miRNA and total viral RNA were isolated from N2a cells and BALB/c mice using Tripure Reagent (Roche, Sweden), and cDNA synthesis was then performed using a Bulge-LoopTM miRNA qRT-PCR Starter Kit (C10211-1, Ribobio, China) for miRNA and a PrimeScript RT Reagent Kit (DRR037A, Takara, Japan) for viral RNA. The RNA samples were tested using an ultraviolet spectrophotometer to verify that there was no DNA contamination. If the OD260/280 value was between 1.8 and 2.0, the sample was used for reverse transcription. The qRT-PCR assay was performed using a SYBR Green Master Mix Kit (Roche, Sweden), and the relative expression of mRNA was analyzed using the 2-△△CT method. We chose the housekeeping genes U6 and GAPDH as internal controls to normalize miR-142a-3p and Rab3a expression, respectively. Bulge-Loop primers for miR-142a-3p (catalog no. C10211-1) and U6 (catalog no. MQPS0000002-1-100) were purchased from RiboBio (Guangzhou, China). The primers for Rab3a, PHEV, and GAPDH were as follows: Rab3a sense, 5’- ATC ATT GGG AAC AGC AGC GT -3’; Rab3a antisense, 5’- TCC TCT TGT CGT TGC GGT AG -3’; PHEV sense, 5’-AGC GAT GAG GCT ATT CCG ACT A-3’; PHEV antisense, 5’- TTG CCA GAA TTG GCT CTA CTA CG -3’; GAPDH sense, 5’- CTC AAC TAC ATG GTC TAC ATG TTC -3’; GAPDH antisense, 5’- ATT TGA TGT TAG TGG GGT CTC GCT C -3’. The cycling conditions for qRT-PCR were as follows: 95°C for 3 min, 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, for a total of 35 cycles.
Dual-luciferase reporter assay
Potential target genes of miR-142a-3p were predicted using online software, including Targetscan Mouse (http://www.targetscan.org/), MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/), and Miranda (http://mirdb.org/). Among the candidate target genes, Rab3a contains a conserved mRNA 3′UTR sequence element that is predicted to be complementary to the seed region of mouse miR-142a-3p. The Rab3a mRNA 3′UTR sequence was amplified from mouse brain by RT-PCR, and the mutation in the miR-142a-3p binding site was amplified using overlap PCR. The PCR-amplified wild-type Rab3a mRNA 3′UTR (Rab3a-WT) and the mutated Rab3a mRNA 3′UTR (Rab3a-MUT) were then cloned into the firefly luciferase pmirGLO reporter (Promega, Madison, USA) using the NheI and XhoI sites. The Dual-Luciferase Reporter Assay System (Promega, Madison, USA), with firefly luciferase (luc2) as the primary reporter was used to monitor the interaction of miR-142a-3p and Rab3a mRNA 3′UTR. Renilla luciferase (hRluc-neo) was used as a control reporter for normalization and selection. HEK293T cells were co-transfected with Rab3a-WT/Rab3a-MUT, miRNA mimic/inhibitor/NC, and the Renilla luciferase construct, and the luciferase activity in these cell lysates was measured at 24 h post-transfection.
Western blot
The selected cells were washed with 0.01 M PBS (pH 7.4), harvested, and lysed on ice for 20-30 min using 100 μL of radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF). Protein was quantified using a BCA Protein Assay Kit (Pierce, USA) according to the manufacturer’s instructions. Protein samples (60 mg per well) were separated by electrophoresis in 12% SDS-polyacrylamide gels and then blotted onto a 0.22-μm polyvinylidene fluoride (PVDF) membrane (Thermo Scientific) using a Bio-Rad wet transfer instrument. The membranes were blocked overnight in blocking buffer and then probed with antibodies against Rab3a (Abcam, 1:2000), β-actin (Abcam, 1:10000), and PHEV (a lab-prepared monoclonal antibody against the PHEV nucleocapsid [N] protein, 1:500) at 4°C overnight. After washing with 0.1% Tween-20/PBS, the membranes were incubated with the corresponding secondary IgG antibody for 1 h at 37°C. The signal was read using an ECL detection kit (Proteintech, USA).
RNA interference and overexpression of Rab3a
The coding sequence (CDS) of Rab3a mRNA was amplified by RT-PCR and cloned into the plasmid eGFP-N1 using the NheI and XhoI sites (Promega, Madison, USA), and the resulting GFP-Rab3a plasmid construct was confirmed by sequencing. The siRNA oligonucleotide 5’- CCA UGG GCU UCA UCC UAA UTT -3’ directed against Rab3a was purchased from GenePharma Co., Ltd (Shanghai, China). Relative mRNA and protein levels were determined using qRT-PCR and Western blotting 48 h after transfection with Rab3a siRNA, GFP-Rab3a, or GFP-N1 (2 μg per well). The primers for the Rab3a CDS were as follows: Rab3a-CDS sense primer, 5’- GCT AGC ATG GCT TCC GCC ACA GAC TC -3’; Rab3a-CDS antisense primer, 5’- CTC GAG GCA GGC ACA ATC CTG ATG AG -3’.
Immunofluorescence analysis
After transfection for indicated times, the cells were harvested and washed with 0.01 M PBS, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.25% Triton X-100 for 15 min, and blocked with blocking buffer for 1 h at 37°C. The cells were then incubated with the indicated antibodies at 4°C for 24-36 h. After washing, the samples were incubated with a secondary antibody conjugated with Alexa Fluor 488 or Alexa Fluor 594 for 1 h at 37°C, and the nuclei were stained with Hochest 33342. Images of the samples were obtained using a confocal microscope.
Images and statistical analysis
GraphPad Prism software was used to produce histograms and for data analysis by t-test or one-way ANOVA. The fold change in band density in Western blot images was calculated using Tanon Gis software (Tanon, Shanghai, China). A P-value of 0.05 was defined as the threshold for statistical significance.
Results
PHEV infection induces upregulation of miR-142a-3p
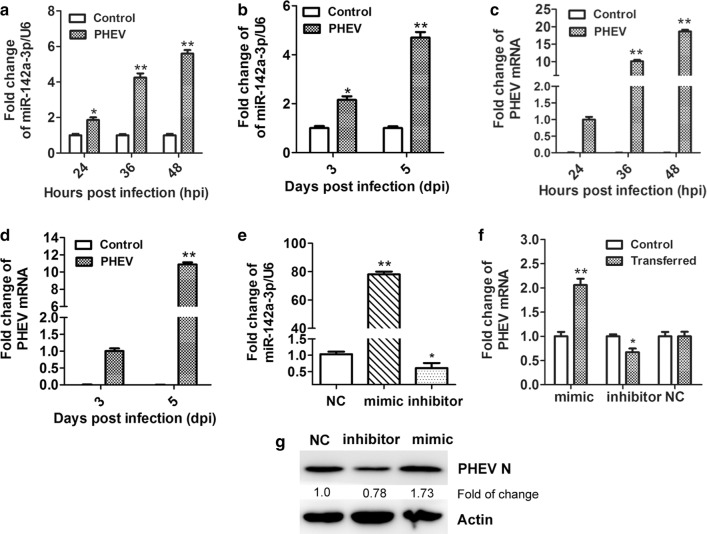
To measure the level of miR-142a-3p during PHEV infection, N2a cells were infected with PHEV at an MOI of 10 and harvested at 24, 36, and 48 hpi. The level of miR-142a-3p RNA was then determined by qRT-PCR, and we found that its relative expression was significantly increased, up to 5.60-fold at 48 hpi (Fig. 1a). Three-week-old male BALB/c mice were inoculated with 100 μL of PHEV (104.45 TCID50/0.1 mL) by the intranasal route, and their brain tissues were harvested at 3 and 5 dpi. As shown in Fig. 1b, the expression of miR-142a-3p RNA was upregulated by 4.70-fold at 5 dpi in PHEV-infected mice. To determine whether change in miR-142a-3p expression correlate with PHEV infection, qRT-PCR assays were used to measure replication of the PHEV genome in infected N2a cells and mice (Fig. 1c and d). The results showed that viral replication increased in a time-dependent manner, suggesting that miR-142a-3p expression during PHEV infection is possibly associated with PHEV proliferation.
Fig. 1.
miR-142a-3p is upregulated during PHEV infection in vitro and in vivo. (a) PHEV-infected N2a cells were collected at different time points. The expression of miR-142a-3p was measured by qRT-PCR. (b) miR-142a-3p expression in mouse brain tissues after infection with PHEV was measured by qRT-PCR. (c) Cells that were pre-treated as indicated above were analyzed by qRT-PCR to measure the level of PHEV mRNA. (d) PHEV mRNA expression in mouse brain tissues after infection with PHEV was measured by qRT-PCR. (e) N2a cells were transfected with miR-142a-3p mimic (100 nM), inhibitor (400 nM), or NC (100 nM). Expression of miR-142a-3p was then measured by qRT-PCR, with the housekeeping gene U6 used as an internal control for normalization. (f) N2a cells were transfected with the miR-142a-3p mimic, inhibitor, or NC for 12 h, followed by PHEV infection for 24 h. Lysates were harvested and tested by qRT-PCR. (g) Cells were pre-treated as indicated above, lysed, and analyzed using a Western blotting assay to determine the level of PHEV N protein. Data were normalized to β-actin. All of the data are representative of at least three independent experiments (*, p < 0.05; **, p < 0.01)
Enhanced function of miR-142a-3p promotes PHEV proliferation
To investigate whether miR-142a-3p affects PHEV proliferation, N2a cells were transfected with chemically synthesized miR-142a-3p mimic (100 nM), inhibitor (400 nM), or negative control (NC). At 12 h post-transfection, the cells were incubated with PHEV for 24 h and then harvested. The RNA levels of miR-142a-3p and PHEV were measured using qRT-PCR, and we found that transfection with the mimic oligonucleotide significantly increased the overall level of miR-142a-3p RNA and facilitated PHEV genome replication (Fig. 1e and f). In contrast, downregulation of miR-142a-3p by its inhibitor led to a suppression of PHEV proliferation, but the effect was small (Fig. 1e and f). PHEV N protein production in these cells was then analyzed by Western blotting. As shown in Fig. 1g, miR-142a-3p promotes the expression of PHEV N protein 48 h after transfection (Fig. 1g). Together, our results suggest that miR-142a-3p positively regulates PHEV proliferation in N2a cells.
Rab3a expression is suppressed in response to PHEV infection
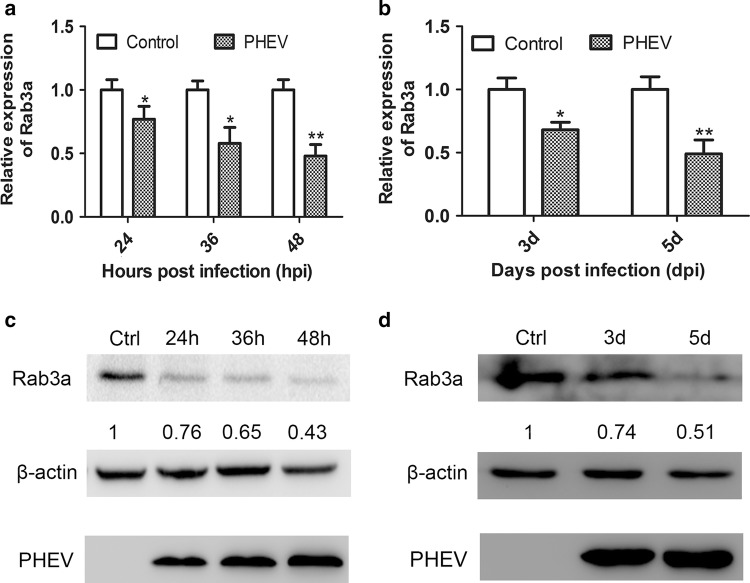
We hypothesized that miR-142a-3p facilitates PHEV replication by regulating oncogenes and genes involved in viral RNA replication or packaging. To identify potential target mRNAs for miR-142a-3p, bioinformatics software, including Targetscan Mouse, MicroCosm Targets, and Miranda, were used. We focused on a set of seven genes (Ptpn23, Rab3a, Rab12, Tmem55b, Stx12, Fyco1, and Rab14) that were shown previously to be downregulated genes in the CNS using an mRNA array [19]. Of these, Rab3a is of particular interest. It is a small RAB GTPase that plays a role in neurotransmitter release and contains conserved 3’UTR sequence elements that are partially complementary to miR-142a-3p [21]. In consideration of its strong association with virus infection and nerve injury, we selected Rab3a for further verification. To confirm the regulation of Rab3a expression in response to PHEV infection, the infected cell lysates were made at 24, 36, and 48 hpi, and mouse brain tissues were harvested at 3 and 5 dpi. Using qRT-PCR and Western blot analysis, we observed an apparent downregulation of Rab3a mRNA and protein expression in N2a cells during PHEV infection in a time-dependent manner (Fig. 2a and c). Similar effects were also detected in PHEV-infected BALB/c mice (Fig. 2b and d). The findings suggest that PHEV negatively regulates expression of endogenous Rab3a.
Fig. 2.
Rab3a expression is inhibited during PHEV infection in vitro and in vivo. (a) N2a cells were mock infected (control) or infected with PHEV for the indicated times. Endogenous Rab3a mRNA expression was measured by qRT-PCR and compared to that of the control. (b) Mice were mock infected (control) or infected with PHEV. At 3 or 5 dpi, the cerebral cortexes were harvested and Rab3a mRNA was quantitated by qRT-PCR and normalized to GAPDH. (c) Lysates of mock-infected N2a cells or cells that were infected with PHEV were analyzed by Western blotting using primary antibodies against Rab3a or β-actin. The data were normalized to β-actin. (d) Lysates from cerebral cortexes of mice that were pre-treated as indicated above were analyzed by Western blotting and normalized to β-actin. All of the data are representative of at least three independent experiments (*, p < 0.05; **, p < 0.01)
Rab3a is a target gene of miR-142a-3p
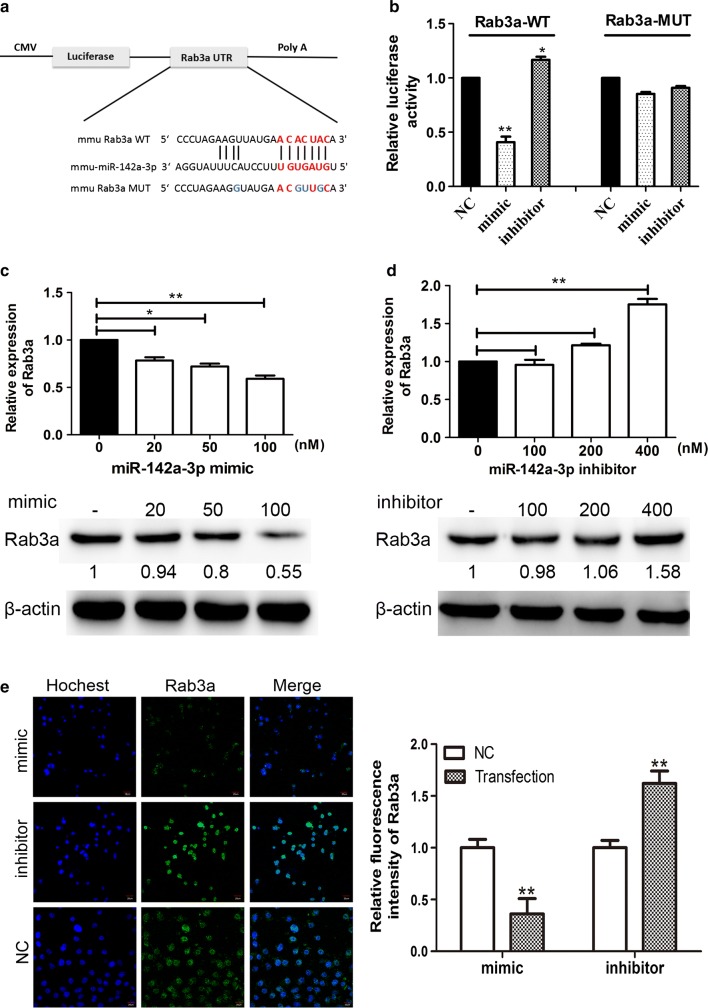
To investigate whether miR-142a-3p is capable of targeting the Rab3a mRNA 3′UTR, luciferase reporter constructs expressing the wild-type (WT) or mutated (MUT) Rab3a 3′UTR were generated (Fig. 3a). A dual-luciferase reporter assay was performed at 24 h after co-transfection of HEK293T cells with miR-142a-3p mimic/inhibitor, Rab3a-WT/Rab3a-MUT luciferase reporter, and Renilla luciferase reporter. We found that the relevant luciferase activity in cells co-transfected with miR-142a-3p mimic and Rab3a-WT was greatly reduced, while mimic- and Rab3a-MUT-overexpressing cells showed no change in luciferase activity (Fig. 3b). Cells that were co-transfected with miR-142a-3p inhibitor and Rab3a-WT plasmid showed a significant increase in luciferase activity compared to the NC and cells overexpressing Rab3a-WT (Fig. 3b). Next, we transfected N2a cells with miR-142a-3p mimic (20, 50, and 100 nM) or inhibitor (100, 200, and 400 nM), in and the lysates were tested by qRT-PCR and Western blotting. We found that the mimic transfection resulted in a dose-dependent reduction of Rab3a expression (Fig. 3c). In contrast, the mRNA and protein level of Rab3a were dose-dependently upregulated in N2a cells after transfection with miR-142a-3p inhibitor (Fig. 3d). Finally, the expression of Rab3a in cells transiently expressing miR-142a-3p mimic (100 nM) or inhibitor (400 nM) was measured by immunofluorescence analysis (Fig. 3e). Representative micrographs and quantitative analysis of the average fluorescence intensity of Rab3a suggested that miR-142a-3p mimic caused a significant reduction in the Rab3a signal, and that the miR-142a-3p inhibitor suppressed the effect (Fig. 3e). These findings suggest that miR-142a-3p binds to the target site in the Rab3a mRNA 3′UTR and negatively regulates its expression in a dose-dependent manner.
Fig. 3.
Rab3a is a target gene of miR-142a-3p. (a) Schematic representation of WT or MUT luciferase reporter of Rab3a mRNA 3′UTR. The frame and letters in blue indicate the point mutant. (b) HEK293T cells were cotransfected with Rab3a-WT or Rab3a-MUT reporter constructs and the indicated oligonucleotides (100 nM mimic, 400 nM inhibitor, or 100 nM NC) for 24 h. The cells were harvested, and Renilla and firefly luciferase activity was measured. (c) The mRNA and protein levels of Rab3a in N2a cells transfected with miR-142a-3p mimic were determined by RT-PCR and Western blotting. (d) The expression of Rab3a was measured in N2a cells after transfection with miR-142a-3p inhibitor. (e) Representative micrograph of immunofluorescence analysis for Rab3a expression (green) in N2a cells transfected with the miR-142a-3p mimic- or miR-142a-3p inhibitor. The cell nucleus was stained with Hochest33342 (blue). Quantitative analysis of the Rab3a staining is shown on the right. The bars indicate 20 μm. All of the data are representative of at least three independent experiments (*, p < 0.05; **, p < 0.01)
miR-142a-3p enhances PHEV proliferation by targeting Rab3a in N2a cells
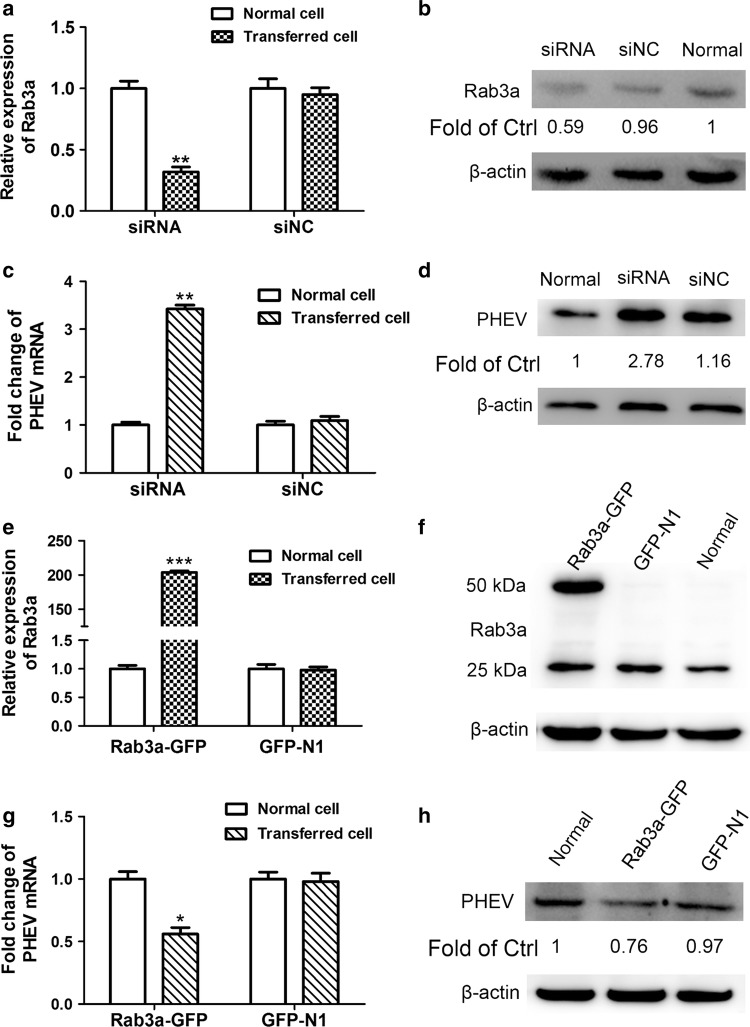
To assess whether Rab3a is important for PHEV proliferation in N2a cells, we performed knockdown and overexpression studies of Rab3a. N2a cells were transfected with siRNA targeting Rab3a (siRab3a) and NC (the disordered base sequence, siNC) for 24 h, and the silencing effect was confirmed (Fig. 4a and b). These pretreated cells were subsequently infected with PHEV at an MOI of 10 for another 24 h, and qRT-PCR and Western blot assays revealed that PHEV proliferation significantly increased when Rab3a was functionally inactivated, compared with NC-transfected cells (Fig. 4c and d). To investigate the effect of a gain of Rab3a function, a GFP-N construct encoding Rab3a CDS (Rab3a-GFP) was generated and used to transiently transfect N2a cells for 24 h. As shown in Fig. 4e and f, increased expression of Rab3a mRNA and protein was detected in the Rab3a-GFP-expressing cells. In the representative blot, the upper band corresponds to overexpressed exogenous Rab3a protein (~50 kDa), while the bottom band is endogenous Rab3a protein (~25 kDa). These cells were subsequently infected with PHEV, and we found that Rab3a overexpression resulted in suppression of viral proliferation at 24 dpi when compared to GFP-transfected cells (Fig. 4g and h). Taken together, our data indicate that miR-142a-3p might upregulate PHEV proliferation by targeting Rab3a, which plays a passive role in viral replication.
Fig. 4.
Rab3a plays a passive regulatory role in PHEV proliferation. (a) The expression of Rab3a mRNA in N2a cells transfected with 50 nM siRab3a or siNC was measured by qRT-PCR. Normal cells were transfected without any siRNA. (b) The protein levels of Rab3a in N2a cells transfected with siRNA were determined by Western blotting. (c) The levels of viral genomic RNA were measured after transfecting with Rab3a siRNA. Untransfected cells were used as a control. (d) Western blotting was performed to examine the expression of PHEV N protein. (e and f) The expression of Rab3a in cells that transfected with Rab3a-GFP was examined by qRT-PCR (e) and Western blotting (f). (g and h) The viral genomic RNA and PHEV N protein levels in Rab3a-GFP-transfected cells were determined. All of the data are representative of at least three independent experiments (*, p < 0.05; **, p < 0.01; ***, p <0.001)
Discussion
miRNAs are ~22-nt ncRNAs that play essential roles in complex virus-host interactions at the posttranscriptional level [22, 23]. The specific target mRNA is principally recognized by the seed region of miRNAs, though multiple validated targets lack perfect base-pairing interactions. miRNA target sequences could be involved in the control of exogenous gene expression and viral tropism in specific cells or tissues, and miRNA-based therapeutics might be designed to either restore or suppress the function of dysregulated miRNAs. Given the versatility and importance of miRNAs, both DNA and RNA viruses affect host pathways by destroying, boosting, or hijacking miRNAs to benefit the viral life cycle [24]. In the case of PHEV, a highly neuropathic virus, little is known about how miRNAs participate in its infection and pathological process. Lv et al. [25] reported that enrichment of miR-21a-5p increases PHEV proliferation by targeting the protein Caskin1, which is required for neuronal axon growth in vivo and in vitro. Here, we attempted to identify miR-142a-3p-mediated molecular mechanisms underlying PHEV pathogenesis.
PHEV is a severely neuropathic virus that invades the CNS of piglets by the age of 3 weeks, which causes typical nerve dysfunction. Previous studies have reported that miR-142 plays a variety of vital roles in various diseases, such as virus infection, inflammation, and cancer [12, 26]. In addition, miR-142 could be a promising biomarker for the diagnosis of many diseases [27]. Thus, the functional significance of miR-142a-3p during PHEV infection is of interest for research and antiviral therapy. It has been shown that miR-142a-5p disrupts neuronal morphogenesis to promote PHEV infection [28]. Although miR-142a-3p and miR-142a-5p originate from the miR-142a hairpin [12], the relationship between miR-142 and PHEV infection is poorly understood. Here, we verified the interaction between PHEV and miR-142a-3p and found that the amount of miR-142a-3p in mice had increased significantly 5 days after infection with PHEV. Meanwhile, miR-142a-3p RNA was also apparently upregulated in PHEV-infected N2a cells, implying that miR-142a-3p might be involved in PHEV infection in vitro and in vivo. Moreover, our data revealed that enrichment of miR-142a-3p could promote PHEV infection or propagation and that knockdown of miR-142a-3p had an opposite effect. We therefore concluded that miR-142a-3p could affect PHEV accumulation and might also play a specific role in PHEV pathogenesis. More follow-up work is needed to support this view.
How might miR-142a-3p play a role during PHEV infection? Sequencing, bioinformatics, and time course experiments indicated that miR-142a-3p directly targets the Rab3a mRNA 3′UTR. Rab3a is a small G protein that belongs to the RAS family of proteins. It regulates vesicle trafficking and is involved in calcium-triggered exocytosis in nerve cells [29, 30]. Generally, Rab3a protein localizes to the Golgi complex and regulates the synthesis and transport of effectors by interacting with GTPase [31]. Given that PHEV buds from endoplasmic reticulum–Golgi intermediate compartments and is assembled within small vesicles in the trans-Golgi network, we hypothesize that Rab3a, a Golgi-associated protein, may acts as a regulator of PHEV replication or nucleocapsid assembly [11, 32]. In this study, we found that Rab3a deficiency promotes PHEV replication and propagation, while overexpression of Rab3a restricts virus proliferation. Thus, we draw the preliminary conclusion that, in some phase of PHEV infection, Rab3a functions as a negative regulator through base-pairing interactions with miR-142a-3p.
In summary, the biological significance of miR-142a-3p-mediated Rab3a suppression and the underlying mechanisms of the response to PHEV infection remain to be elucidated. Although some host miRNAs execute antiviral functions by limiting viral infection in cells, it is still debatable whether an miR-142a-3p antagonist could be used as a potential antiviral therapy against PHEV infection in the physiological context. Much more work in this area still needs to be done in the future.
Acknowledgements
Peng Fan, Zi Li, and Wenqi He designed and supervised the experiments. Peng Fan and Jiyu Guan performed most of the tests. Xiaoling Lv and Fang Li cultured cells and analyzed immunofluorescence images. Shiyu Hu and Yungang Lan constructed plasmids. Hongbin He and Gencheng Fan prepared the figures. Kui Zhao drafted the manuscript, and Feng Gao revised it for valuable intellectual content.
Funding
This study was supported by the National Key Research and Development Program of China (Grant 2016YFD0500102), the National Natural Science Foundation of China (Grants 31902262, 31872446, 31772704, 31672519, and 31602018), the State Key Laboratory of Genetically Engineered Veterinary Vaccines (no. AGVSKL-ZD-201808), the China Postdoctoral Science Foundation (Grant 2018M640285).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peng Fan and Jiyu Guan contributed equally to this article.
References
- 1.Dong B, Lu H, Zhao K, Liu W, Gao W, Lan Y, Zhao J, Tang B, Song D, He W, Gao F. Identification and genetic characterization of porcine hemagglutinating encephalomyelitis virus from domestic piglets in China. Arch Virol. 2014;159:2329–2337. doi: 10.1007/s00705-014-2070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao W, Zhao K, Zhao C, Du C, Ren W, Song D, Lu H, Chen K, Li Z, Lan Y, Xie S, He W, Gao F. Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin, China. Virol J. 2011;8:130. doi: 10.1186/1743-422X-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roe CK, Alexander TJ. A disease of nursing pigs previously unreported in Ontario. Can J Comp Med Vet Sci. 1958;22:305–307. [PMC free article] [PubMed] [Google Scholar]
- 4.Greig AS, Mitchell D, Corner AH, Bannister GL, Meads EB, Julian RJ. A Hemagglutinating Virus Producing Encephalomyelitis in Baby Pigs. Can J Comp Med Vet Sci. 1962;26:49–56. [PMC free article] [PubMed] [Google Scholar]
- 5.Lan Y, Zhao K, Wang G, Dong B, Zhao J, Tang B, Lu H, Gao W, Chang L, Jin Z, Gao F, He W. Porcine hemagglutinating encephalomyelitis virus induces apoptosis in a porcine kidney cell line via caspase-dependent pathways. Virus Res. 2013;176:292–297. doi: 10.1016/j.virusres.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alsop JE. A presumptive case of vomiting and wasting disease in a swine nucleus herd. J Swine Health Prod. 2006;14:97–100. [Google Scholar]
- 7.Quiroga MA, Cappuccio J, Pineyro P, Basso W, More G, Kienast M, Schonfeld S, Cancer JL, Arauz S, Pintos ME, Nanni M, Machuca M, Hirano N, Perfumo CJ. Hemagglutinating encephalomyelitis coronavirus infection in pigs, Argentina. Emerg Infect Dis. 2008;14:484–486. doi: 10.3201/eid1403.070825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, He WQ, Lan YG, Zhao K, Lv XL, Lu HJ, Ding N, Zhang J, Shi JC, Shan CJ, Gao F. The evidence of porcine hemagglutinating encephalomyelitis virus induced nonsuppurative encephalitis as the cause of death in piglets. PeerJ. 2016;4:e2443. doi: 10.7717/peerj.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorbach JN, Wang LY, Nolting JM, Benjamin MG, Killian ML, Zhang Y, Bowman AS. Porcine hemagglutinating encephalomyelitis virus and respiratory disease in exhibition swine, Michigan, USA, 2015. Emerg Infect Dis. 2017;23:1168–1171. doi: 10.3201/eid2307.170019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirano N, Nomura R, Tawara T, Tohyama K. Neurotropism of swine haemagglutinating encephalomyelitis virus (coronavirus) in mice depending upon host age and route of infection. J Comp Pathol. 2004;130:58–65. doi: 10.1016/S0021-9975(03)00083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YC, Bai WZ, Hirano N, Hayashida T, Hashikawa T. Coronavirus infection of rat dorsal root ganglia: ultrastructural characterization of viral replication, transfer, and the early response of satellite cells. Virus Res. 2012;163:628–635. doi: 10.1016/j.virusres.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha A, Carraro G, El Agha E, Mukhametshina R, Chao CM, Rizvanov A, Barreto G, Bellusci S. Generation and validation of miR-142 knock out mice. Plos One. 2015;10:e0136913. doi: 10.1371/journal.pone.0136913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai FW, Stephenson KB, Mahony J, Lichty BD. Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-kappaB activation. J Virol. 2014;88:54–65. doi: 10.1128/JVI.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao XM, Song XJ, Bai XY, Fei NJ, Huang Y, Zhao ZM, Du Q, Zhang HL, Zhang L, Tong DW. miR-27b attenuates apoptosis induced by transmissible gastroenteritis virus (TGEV) infection via targeting runt-related transcription factor 1 (RUNX1) PeerJ. 2016;4:e1635. doi: 10.7717/peerj.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. Plos One. 2009;4:e1635. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 17.Trobaugh DW, Gardner CL, Sun CQ, Haddow AD, Wang EY, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan HB, Liu YJ, Wang L, Du TT, Dong M, Gao L, Meng ZZ, Jin Y, Chen Y, Deng M, Yang HT, Jing Q, Gu AH, Liu TX, Zhou Y. miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood. 2014;124:1320–1330. doi: 10.1182/blood-2013-12-545012. [DOI] [PubMed] [Google Scholar]
- 19.Lan YG, Zhao K, Zhao JK, Lv XL, Wang GL, Lu HJ, Tang B, Li Z, Chang LZ, Jin Z, He WQ, Gao F. Gene-expression patterns in the cerebral cortex of mice infected with porcine haemagglutinating encephalomyelitis virus detected using microarray. J Gen Virol. 2014;95:2192–2203. doi: 10.1099/vir.0.066845-0. [DOI] [PubMed] [Google Scholar]
- 20.Khanna M, Saini S, Shariff M, Ronsard L, Singh JK, Kumar H. Data highlighting miR-155 and GAPDH correlation. Data Brief. 2019;24:103945. doi: 10.1016/j.dib.2019.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman WL, Bill CA, Bykhovskaia M. Rab3a deletion reduces vesicle docking and transmitter release at the mouse diaphragm synapse. Neuroscience. 2007;148:1–6. doi: 10.1016/j.neuroscience.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther. 2015;5:17–36. doi: 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 24.Powdrill MH, Desrochers GF, Singaravelu R, Pezacki JP. The role of microRNAs in metabolic interactions between viruses and their hosts. Curr Opin Virol. 2016;19:71–76. doi: 10.1016/j.coviro.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Lv XL, Zhao K, Lan YG, Li Z, Ding N, Su JJ, Lu HJ, Song DG, Gao F, He WQ. miR-21a-5p contributes to porcine hemagglutinating encephalomyelitis virus proliferation via targeting CASK-interactive protein1 in vivo and vitro. Front Microbiol. 2017;8:304. doi: 10.3389/fmicb.2017.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han B, Zhang Y, Zhang YH, Bai Y, Chen XF, Huang RR, Wu FF, Leng S, Chao J, Zhang JH, Hu G, Yao HH. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Lan YG, Zhao K, Lv XL, Ding N, Lu HJ, Zhang J, Yue HQ, Shi JC, Song DG, Gao F, He WQ. miR-142-5p disrupts neuronal morphogenesis underlying porcine hemagglutinating encephalomyelitis virus infection by targeting Ulk1. Front Cell Infect Microbiol. 2017;7:155. doi: 10.3389/fcimb.2017.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geppert M, Bolshakov VY, Siegelbaum SA, Takei K, De Camilli P, Hammer RE, Sudhof TC. The role of Rab3A in neurotransmitter release. Nature. 1994;369:493–497. doi: 10.1038/369493a0. [DOI] [PubMed] [Google Scholar]
- 30.Geppert M, Goda Y, Stevens CF, Sudhof TC. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature. 1997;387:810–814. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- 31.Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
- 32.Li YC, Bai WZ, Hirano N, Hayashida T, Taniguchi T, Sugita Y, Tohyama K, Hashikawa T. Neurotropic virus tracing suggests a membranous-coating-mediated mechanism for transsynaptic communication. J Comp Neurol. 2013;521:203–212. doi: 10.1002/cne.23171. [DOI] [PMC free article] [PubMed] [Google Scholar]