Abstract
Rotavirus is a non-enveloped virus that depends on cellular lipids for cell entry and associates with lipid rafts during assembly. However, the effects of cellular lipids on rotavirus assembly are still not fully understood. The present study analyzes the effects of lovastatin, an inhibitor of cholesterol biosynthesis, during rotavirus infection in MA104 cells with regard to viral growth and particle assembly. Following viral infection, a 2-log relative reduction of viral titers was observed in drug-treated cells, while viral mRNA levels in infected cells remained unaltered in both groups. Furthermore, the levels of some viral proteins in drug-treated cells were elevated. The observed discordance between the viral RNA and protein levels and the decrease in infectivity titers of viral progeny in the drug-treated cells suggested that the drug affects viral assembly, the viral proteins not being properly incorporated into virions. Transmission electron microscopic (TEM) analysis revealed that in drug-treated cells there was an increase in “empty-looking” rotavirus particles devoid of an electron-dense core as compared to the normal, electron-dense particles seen in untreated infected cells. The present study thus provides visual evidence of defective rotavirus particle assembly as a result of cholesterol depletion.
Keywords: Lovastatin, Lipid Raft, Virus Assembly, MA104 Cell, Cholesterol Depletion
Introduction
Cellular lipids are known to play an important role in the life cycle of many enveloped viruses. As lipids are major constituents of the cell membrane, it is not surprising that they are implicated in cell entry mechanisms of many viruses such as the human immunodeficiency virus (HIV), Epstein-Barr virus (EBV), coronavirus, and the Semliki Forest virus (SFV), to name a few [14, 19, 20, 35]. Besides being involved in viral entry at the membrane level, cellular lipids are also essential for many intracellular processes of viral proteins such as post-translational fatty acid modifications (e.g., acylation, myristylation, palmitoylation and prenylation) and also play an important role in enveloped virus assembly and protein trafficking [1, 10, 15, 34].
As lipids are critical to the biology of enveloped viruses, some of these viruses modulate the host-cell lipid metabolism to their advantage, by up- or down-regulating their lipid synthesis [2, 8, 16, 28]. However, recent reports have demonstrated the involvement of lipids in the cell biology of a non-enveloped, double-stranded RNA virus, the rotavirus, as well [3, 5, 6, 13, 33]. Rotavirus belongs to the family Reoviridae and is the leading etiological agent causing viral gastroenteritis in humans and animals [18, 31]. Parashar et al. [32] estimated a mean of 611,000 (range 454,000–705,000) deaths from rotavirus disease worldwide per annum. The role of lipids in rotaviral entry, specific lipid binding of the viral proteins, association of infectious virions or rotaviral proteins within lipid rafts for assembly, and release have all been documented [3–6, 13, 17, 29, 33]. Noteworthy among these studies are the ones that clearly underlined the role of lipid rafts in rotavirus biology by demonstrating that the viral proteins as well as the triple-layered infectious particle associate with rafts, both in culture and in cells of experimentally infected animals, and that the assembly of VP4 into the viral particle occurs as an extra-ER event [3–6, 33]. Recently Delmas et al. [6] reported distinct differences in raft-type membrane microdomains between Caco-2 cells and another cell line, MA104 (monkey kidney epithelial cells), the two most commonly used cell lines for studying rotavirus replication in vitro. The present study demonstrates the effect of cholesterol depletion on rotavirus titers and assembly in MA104 cells by utilizing lovastatin, which inhibits HMG CoA reductase, the rate-limiting enzyme in cholesterol biosynthesis. Our analysis revealed that while the drug treatment apparently reduces rotavirus titers in MA104 cells, transmission electron microscopy showed that the cholesterol-lowering drug has a profound effect on virus assembly and results in accumulation of defective, “empty-looking” particles inside the cell.
Materials and methods
Cells, virus and lipid-lowering drugs
MA104 cells (ATCC, Manassas, VA) were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech, Herndon, VA) containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), 2 mM L-glutamine (Mediatech, Herndon, VA), 50 IU/ml penicillin and 50 μg/ml streptomycin (Mediatech Inc., Herndon, VA). Simian (SA11) rotavirus (ATCC, Manassas, VA) was propagated in MA104 cells as described previously [21, 22].
Lovastatin treatment, cholesterol estimation and virus infection
A commercially available lovastatin, mevinolin, (Sigma chemicals, St Louis, MO) was used in the present study, and a 100 mM stock solution of the drug was prepared in ethanol and activated as described previously [27]. Prior to infection, MA104 cells were left untreated or treated with 15 μM of the drug, initially in complete DMEM medium for 24 h, followed by an additional 12 h incubation with 15 μM mevinolin constituted in serum-free medium. Virus infection was carried out as described previously [3, 24]. Briefly, SA11 virus was activated with 10 μg/ml of type-V acetylated trypsin, derived from bovine pancreas (Sigma Chemicals, St Louis, MO), for 45 min at 37°C and then overlaid on confluent MA104 cells for 90 min at 37°C. Following initial virus binding, the inoculum was removed, and fresh serum-free medium containing 15 μM mevinolin was added and incubated for 48–72 h. Trypsin was not included in the serum-free medium so as to enable a moderately slow infection such that a greater proportion of the cell monolayer is recovered after 72 h. Cell viability between the two groups was estimated using the trypan blue dye exclusion method. Cellular cholesterol content was estimated using a commercially available colorimetric kit as per the manufacturer’s instructions (Roche, Indianapolis, USA).
Virus titration
Viral titers were estimated by a fluorescent focus assay (FFA) enumerating the number of fluorescent cell focus units (FCFU) per well, as described previously [3, 7]. Briefly, rotavirus-infected cells in triplicate were freeze–thawed and clarified by centrifugation at 3,000×g for 5 min. Following initial virus binding, the inoculum was removed, fresh serum-free medium was added, and incubation was continued for 12 h for FFA. Cells were washed with phosphate-buffered saline (PBS) and fixed in ice-cold acetone for 10 min. Infected cells were immuno-stained with a guinea pig anti-rotavirus NSP5 antibody [12, 23] and detected by secondary incubation with goat anti-guinea-pig FITC-labeled antibody (Vector, Burlingame, CA). Slides were mounted with an anti-fade mounting medium (Vector, Burlingame, CA), and foci of infected cells were counted under an inverted Nikon fluorescent microscope (Model: Eclipse TE2000-U, Nikon, USA).
Immunoblot assay
Mock- and rotavirus-infected MA104 cells were washed with PBS, and lysates were prepared in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 7.2) as per a previous protocol [26]. The protein concentration in lysates was estimated using a commercially available kit based on the Bradford assay (BioRad Labs, Hercules, CA), and a sample volume equivalent to 20 μg each was loaded and resolved by sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis [23–26]. The proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, USA), and the viral proteins were detected using antibodies against rotavirus VP4 [30] or NSP2 proteins [12]. Incubation with a secondary, horseradish-peroxidase-labeled, anti-species IgG antibody (Chemicon, Temecula, CA) was followed by enhanced chemiluminescence signal detection (Pierce, USA). Immunoblots were also probed with a rabbit polyclonal anti-catalase antibody (BioMol, Plymouth Meeting, PA) to detect cellular catalase levels as a “housekeeping marker” to ensure equal protein loading in each lane.
Viral RNA estimation in infected cells and purified virus particles
RT-PCR was performed on RNA extracted from viral particles as well as from infected cells as per a previous protocol [22, 23]. Briefly, duplicate sets of flasks containing MA104 cells, treated with or without lovastatin, were infected with SA11 rotavirus. Following infection, one set of flasks was used for extracting RNA from the infected cells, whereas the other set was used for first isolating the virus particles from infected cells and then extracting RNA from the purified particles. Virus particles were extracted from infected cells by repeated freeze–thawing and then clarified by centrifugation at 3,000×g for 30 min. Virus particles in the supernatant were pelleted by centrifugation through a 40% sucrose cushion in a Beckman SW55 rotor for 90 min at 25,000×g. RNA was extracted using RNA-Stat (Tel-Test Inc., Friendswood, TX) as described previously (22, 23). RT-PCR was performed to detect the levels of gene segments coding for one of the rotavirus structural proteins, VP7, and a non-structural protein, NSP4, using appropriate pairs of primers. RNA levels were semi-quantitated by the end-point dilution method. The VP7 RNA segment was amplified using a set of forward (5′-CGCGGATCCGGCTTTAAAGAGAGAATTTCC-3′) and reverse (5′-GATCTCGAGGGTCACATCGAACAATTCTAA-3′) primers, whereas the NSP4 gene was amplified using a set of previously published primers for SA11 [25].
Transmission electron microscopy (TEM)
TEM analysis was performed on rotavirus-infected cells to analyze the effects of the lipid-lowering drug on rotavirus assembly by visualizing the particle morphology as previously described [24]. Briefly, virus-infected MA104 cells with or without 15 μM mevinolin were incubated for 48 h, washed with PBS and then trypsinized. Cells were pelleted and fixed for 2–3 h with 2% glutaraldehyde-2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. Samples were subsequently stored in PBS at 4°C until further processing. To this end, cells were post-fixed with 2% osmium tetroxide, dehydrated with graded alcohols, and embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and were examined with a Zeiss EM 912 Omega electron microscope [24].
Results
Reduction of rotavirus titer in lovastatin-treated MA104 cells
MA104 cells treated with or without 15 μM lovastatin for at least 36 h prior to infection and for another 72 h post-infection (hpi) were tested for cell viability. As reported earlier, trypan blue staining did not reveal any significant difference in cell viability between drug-treated and untreated MA104 cells [9]. Cellular cholesterol analysis revealed that lovastatin-treated cells undergo a 35–45% reduction in cholesterol levels when compared to normal MA104 cells. Once it was established that the drug was producing its desired cholesterol-reducing effect at 15 μM concentration without significantly affecting cell viability, rotaviral titers at the various time points were estimated by FFA. Time-course analysis revealed that rotaviral titers were comparable between the normal and drug-treated cells at early time points between 12 and 24 h, suggesting no reduction in initial infectivity due to lovastatin treatment (Fig. 1). However, by 48–72 h, a reduction in viral titers of more than two logs was observed in lovastatin-treated cells when compared to normal cells (Fig. 1).
Fig. 1.
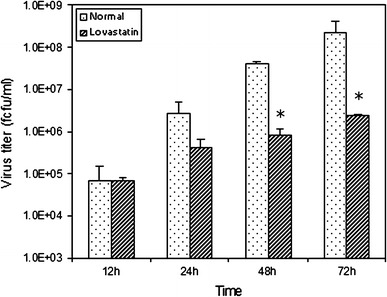
Effect of a lipid-lowering drug on rotavirus infection. Following lovastatin treatment (see “Materials and methods”), cells were infected and samples collected at 12, 24, 48 and 72 h post-infection (hpi). Virus was extracted by repeated freeze–thawing, and viral titers were analyzed by fluorescent focus assay (FFA). As shown in the figure, viral titers in the lovastatin-treated group are quite similar to the control group in the initial phase of infection (12–24 h). However, by 48 and 72 h there is a drastic reduction, greater than a 2-log difference (P < 0.05), in viral titers in the drug-treated group compared to the control group
Lovastatin-treated cells demonstrate increased levels of rotavirus proteins
To analyze whether the lovastatin-induced reduction in viral titers was due to a reduction in viral translation, viral protein levels were compared between rotavirus-infected normal and lovastatin-treated MA104 cells. Immunoblot analysis revealed that the viral protein levels, represented by VP4 and NSP2, were unchanged until the earlier time point of 24 hpi. However, by 48 hpi the VP4 and NSP2 levels were increased multifold compared to protein levels in normal rotavirus-infected cells (Fig. 2). The levels of catalase protein were uniform in all wells, indicating an equal protein concentration in all lanes (Fig. 2).
Fig. 2.
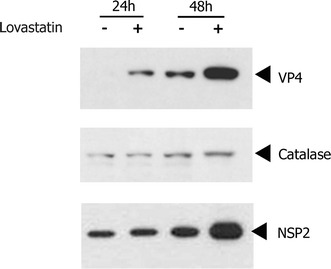
Effect of lovastatin on viral protein levels. Infected-cell lysates from drug-treated and control cells were analyzed to demonstrate viral protein levels by immunoblot assay. Rotavirus antibodies against a structural (VP4) and a non-structural (NSP2) protein were used to detect viral protein levels in both cell lysates. Increased levels of both VP4 and NSP2 proteins are observed in the statin-treated cells by 48 hpi. Cellular catalase levels, detected by using a rabbit anti-catalase antibody, are indicated to demonstrate equal protein loading in all of the lanes
Lovastatin-treated cells exhibit similar levels of viral mRNA as untreated cells
The RT-PCR analysis indicated that the mRNA levels of gene segment 9 (VP7) within the infected cells remained unchanged between the drug-treated and normal MA104 cells at 72 hpi (Fig. 3). However, when mRNA of gene 9 from purified particles was subjected to RT-PCR, the results indicated a substantial decrease in viral RNA levels in particles derived from the drug-treated cells. To ensure reproducibility, mRNA levels of an additional rotavirus gene, segment 10 (NSP4), was tested as well by RT-PCR. Since results from the gene 10 (NSP4) and gene 9 (VP7) levels were identical in the RT-PCR analysis, only VP7 findings are presented as a representation of the analysis (Fig. 3).
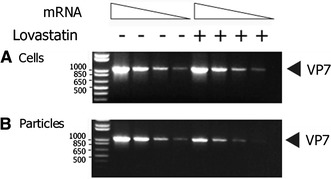
Lovastatin-treated infected cells produce “empty-looking” rotavirus particles
Given our results so far that free viral proteins were abundantly synthesized in the drug-treated cells, while the virus infectivity titers were decreased, we hypothesized that proper viral assembly is affected by the drug treatment. To test this hypothesis, we performed transmission electron microscopic analysis of rotavirus-infected MA104 cells treated with or without lovastatin treatment. The analysis indicated that untreated MA104 cells produce, to a large extent, normal virus particles with an electron-dense core (Fig. 4). However, the drug-treated cells had a relatively much lower number of normal virus particles but with an increased number of “empty-looking” particles that lack a characteristic electron-dense core (Fig. 5), suggesting a defect in virus assembly. These empty particles were uniformly sized and were closer in size to the triple-layered rotavirus particle (minus the VP4 spikes), ruling out the possibility that these empty structures could be cellular vesicles. Although previous reports have demonstrated that lipid depletion affects rotaviral protein targeting and particle assembly, this has not bee supported by in situ visual evidence.
Fig. 4.
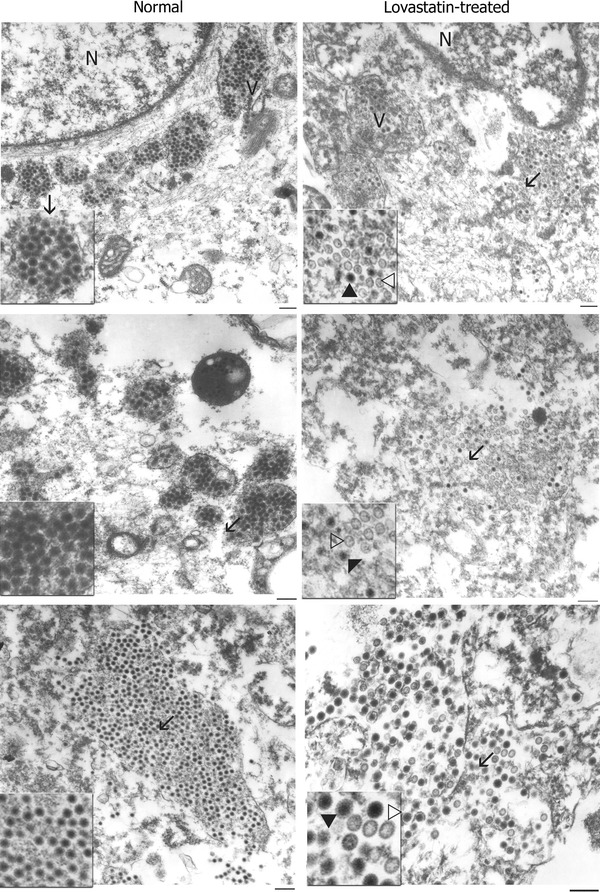
In situ analysis of rotavirus infection at 48 hpi in normal and drug-treated cells by transmission electron microscopy (TEM). Rotavirus-infected cells were processed as described in “Materials and methods” and analyzed by TEM. Three different representative micrographs each of normal and drug-treated MA104 cells infected with rotavirus SA11 are indicated here. Normal MA104 cells infected with rotavirus contain characteristic normal rotavirus particles that contain an electron-dense core, as indicated by filled arrows. A small proportion of the particles within the normal cells lack the electron-dense core and appear as empty particles, as indicated by open arrows. In statin-treated cells, an increase was observed in the number of ‘empty-looking’ particles that exhibit the lack of an electron-dense core (indicated by open arrows). A specific area of the micrograph (indicated by a line arrow) is magnified as an inset image for greater resolution. N nucleus, V viroplasm. Magnification bar represents 300 nm
Fig. 5.
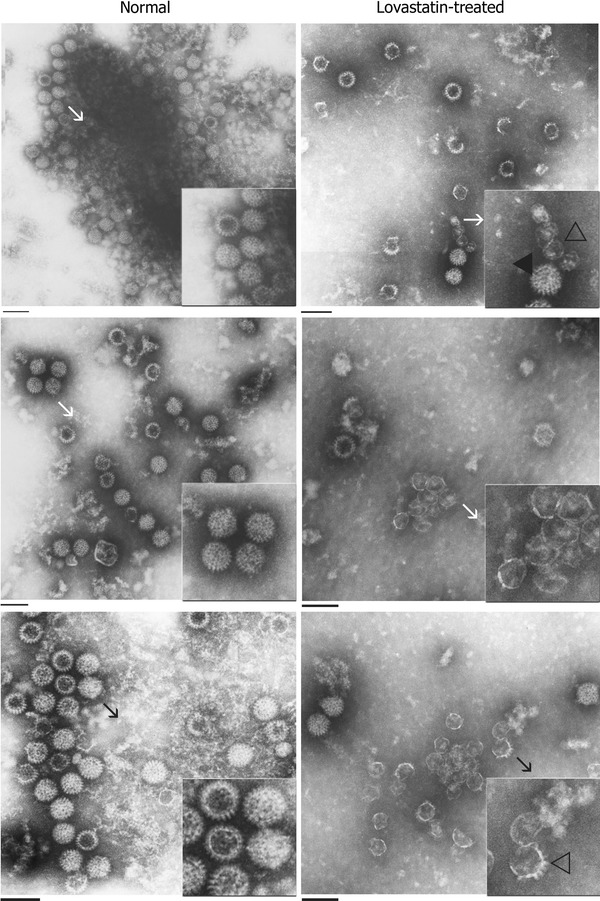
TEM analysis of purified virus from normal and drug-treated cells at 48 hpi. Virus was extracted as described in “Materials and methods” and subjected to TEM analysis. Three different representative micrographs each of virus extracted from normal and drug-treated MA104 cells are indicated here. Virus extracts purified from normal MA104 cells contain more characteristic normal rotavirus particles that contain an electron-dense core (indicated by filled arrows). Virus material extracted from drug-treated cells demonstrates an increase in the number of ‘empty-looking’ particles that lack an electron-dense core (indicated by open arrows). A specific area of the micrograph (indicated by a line arrow) is magnified as an inset image for greater resolution. Magnification bar represents 100 nm
In order to gain a quantitative estimate of the observed difference in virus assembly at 48 hpi between the drug-treated and control groups, electron micrographs of areas containing both populations of particles were taken at random per cell. Micrographs were printed at a magnification of approximately 60,000× and coded to blind the study, and the viral particles were counted and classified. A total of 1,663 particles were counted in 6 micrographs of normal cells. Of these, 1,392 (84%) had a dense core and 271 (16%) lacked the dense core. In the lovastatin-treated cells, out of a total of 1016 particles counted from 6 micrographs, 641 (63%) had dense cores and 375 (37%) were “empty-looking” particles lacking the electron-dense core. This clearly demonstrated that lovastatin-treated cells contained a higher proportion of aberrant virus particles than the control group.
Discussion
The present study demonstrates that cholesterol depletion by lovastatin treatment results in defective rotavirus particle assembly in MA104 cells. The concentration of lovastatin (15 μM) used in the present study is in concurrence with previously published reports in which results from a trypan blue cell viability assay revealed no significant difference in cell viability between normal and the drug-treated cells, suggesting that the drug does not adversely affect cell viability at the concentration used [9].
It is well established that lovastatin reduces intracellular cholesterol levels and that cellular cholesterol is required for rotavirus infectivity and assembly [3–6, 9, 13, 33]. However, so far there have been no reports suggesting a direct effect of lovastatin on viral protein levels in the infected cell. In this report, to investigate whether there was any difference in the availability of viral structural proteins and the non-structural proteins for virus assembly following lovastatin treatment, we selected VP4, which represents a viral structural protein and NSP2, as a representative non-structural protein, to measure viral protein levels. A reduction in the levels of either of these proteins would have indicated a direct effect on virus assembly and infectivity. However, the present analysis revealed an increase in viral protein levels following lovastatin treatment, which was unexpected and suggested that although the viral proteins were readily available in abundance, due to a mechanism that has not yet been identified, rotaviral assembly was being negatively affected. A very similar effect had been reported previously with Rous sarcoma virus infectivity in cells treated with another lipid-reducing agent, cerulenin [11]. However, in this case too, the underlying mechanism is not known.
Previous studies have demonstrated an association of triple-layered infectious rotavirus virions with cellular membrane-associated lipid rafts and the loss of such association following treatment of the cells with a lipid-lowering drug [3–6, 17, 33]. So far, all of these reports have demonstrated the biochemical role of cholesterol in viral entry and association of viral proteins and infectious virions with lipid rafts, but none have conclusively provided visual demonstration of the effect of lipid-lowering drugs on virion assembly. In contrast, the current study presents visual evidence of the effects of cellular lipid-lowering, drug-mediated perturbations in rotavirus biology related to virus assembly.
Acknowledgments
The authors wish to thank Drs. Harry Greenberg, Stanford University, CA, and Oscar Burrone, ICGEB, Trieste, Italy for their generous gift of rotavirus VP4, NSP5 and NSP2 antisera respectively. The technical assistance of Marilyn Lundquist for EM sample preparation is gratefully acknowledged. Critical review of the manuscript by Drs. Stephen Feinstone and Zhiping Ye of CBER is duly acknowledged.
Footnotes
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination policy.
References
- 1.Ahola T, Kujala P, Tuittila M, Blom T, Laakkonen P, Hinkkanen A, Auvinen P. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J Virol. 2000;74:6725–6733. doi: 10.1128/JVI.74.15.6725-6733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter JK, Smith RE. Specificity of avian leukosis virus-induced hyperlipidemia. J Virol. 1984;50:301–308. doi: 10.1128/jvi.50.2.301-308.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuadras MA, Greenberg HB. Rotavirus infectious particles use lipid rafts during replication for transport to the cell surface in vitro and in vivo. Virology. 2003;313:308–321. doi: 10.1016/S0042-6822(03)00326-X. [DOI] [PubMed] [Google Scholar]
- 4.Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J Virol. 2006;80:3935–3946. doi: 10.1128/JVI.80.8.3935-3946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmas O, Gardet A, Chwetzoff S, Breton M, Cohen J, Colard O, Sapin C, Trugnan G. Different ways to reach the top of a cell. Analysis of rotavirus assembly and targeting in human intestinal cells reveals an original raft-dependent, Golgi-independent apical targeting pathway. Virology. 2004;327:157–161. doi: 10.1016/j.virol.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 6.Delmas O, Breton M, Sapin C, Le Bivic A, Colard O, Trugnan G. Heterogeneity of raft-type membrane microdomains associated with VP4, the rotavirus spike protein, in Caco-2 and MA104 cells. J Virol. 2007;81:1610–1618. doi: 10.1128/JVI.01433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes MK, Graham DY. Identification of rotaviruses of different origins by the plaque-reduction test. Am J Vet Res. 1980;41:151–152. [PubMed] [Google Scholar]
- 8.Etingin OR, Hajjar DP. Evidence for cytokine regulation of cholesterol metabolism in herpesvirus-infected arterial cells by the lipoxygenase pathway. J Lipid Res. 1990;31:299–305. [PubMed] [Google Scholar]
- 9.Falke P, Mattiasson I, Stavenow L, Hood B. Effects of a competitive inhibitor (mevinolin) of 3-hydroxy-3-methylglutaryl coenzyme A reductase on human and bovine endothelial cells, fibroblasts and smooth muscle cells in vitro. Pharmacol Toxicol. 1989;64:173–176. doi: 10.1111/j.1600-0773.1989.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 10.Furuishi K, Matsuoka H, Takama M, Takahashi I, Misumi S, Shoji S. Blockage of N-myristoylation of HIV-1 gag induces the production of impotent progeny virus. Biochem Biophys Res Commun. 1997;237:504–511. doi: 10.1006/bbrc.1997.7178. [DOI] [PubMed] [Google Scholar]
- 11.Goldfine H, Harley JB, Wyke JA. Effects of inhibitors of lipid synthesis on the replication of Rous sarcoma virus: A specific effect of cerulenin on the processing of major non-glycosylated viral structural proteins. Biochim Biophys Acta. 1978;512:229–240. doi: 10.1016/0005-2736(78)90249-3. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez SA, Burrone OR. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology. 1991;182:8–16. doi: 10.1016/0042-6822(91)90642-O. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero CA, Zarate S, Corkidi G, Lopez S, Arias CF. Biochemical characterization of rotavirus receptors in MA104 cells. J Virol. 2000;74:9362–9371. doi: 10.1128/JVI.74.20.9362-9371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyader M, Kiyokawa E, Abrami L, Turelli P, Trono D. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J Virol. 2002;76:10356–10364. doi: 10.1128/JVI.76.20.10356-10364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain M, Moss B. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J Virol. 2003;77:9008–9019. doi: 10.1128/JVI.77.16.9008-9019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilback NG, Mohammed A, Fohlman J, Friman G. Cardiovascular lipid accumulation with Coxsackie B virus infection in mice. Am J Pathol. 1990;136:159–167. [PMC free article] [PubMed] [Google Scholar]
- 17.Ishaceka P, Realpe M, Romero P, Lopez S, Arias CF. Rotavirus RRV associates with lipid membrane microdomains during cell entry. Virology. 2004;322:370–381. doi: 10.1016/j.virol.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Kapikian AZ, Hoshino Y, Chanock RM. Rotaviruses. In: Fields BN, Knipe BN, Howley PM, editors. Fields Virology. 3rd. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1787–1833. [Google Scholar]
- 19.Katzman RB, Longnecker R. Cholesterol-dependent infection of Burkitt’s lymphoma cell lines by Epstein-Barr virus. J Gen Virol. 2003;84:2987–2992. doi: 10.1099/vir.0.19252-0. [DOI] [PubMed] [Google Scholar]
- 20.Lu YE, Kielian M. Semliki forest virus budding: assay, mechanisms, and cholesterol requirement. J Virol. 2000;74:7708–7719. doi: 10.1128/JVI.74.17.7708-7719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan KV, Atreya CD. Comparative sequence analysis identified mutations outside the NSP4 cytotoxic domain of tissue culture-adapted ATCC-Wa strain of human rotavirus and a novel inter-species variable domain in its C-terminus. Arch Virol. 2000;145:1789–1799. doi: 10.1007/s007050070056. [DOI] [PubMed] [Google Scholar]
- 22.Mohan KV, Dermody TS, Atreya CD. Mutations selected in rotavirus enterotoxin NSP4 depend on the context of its expression. Virology. 2000;275:125–132. doi: 10.1006/viro.2000.0484. [DOI] [PubMed] [Google Scholar]
- 23.Mohan KV, Som I, Atreya CD. Identification of a peroxisomal targeting signal 1(PTS1)-containing viral protein and its targeting to peroxisomes. J Virol. 2002;76:2543–2547. doi: 10.1128/jvi.76.5.2543-2547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohan KV, Muller J, Atreya CD. The N- and C-terminal regions of rotavirus NSP5 are the critical determinants for the formation of viroplasm-like structures independent of NSP2. J Virol. 2003;77:12184–12192. doi: 10.1128/JVI.77.22.12184-12192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan KV, Kulkarni S, Glass RI, Zhisheng B, Atreya CD. A human vaccine strain of lamb rotavirus (Chinese) NSP4 gene: complete nucleotide sequence and phylogenetic analyses. Virus Genes. 2003;26:185–192. doi: 10.1023/A:1023491514820. [DOI] [PubMed] [Google Scholar]
- 26.Mohan KV, Glass RI, Atreya CD. Comparative molecular characterization of gene segment 11-derived NSP6 from lamb rotavirus LLR strain used as a human vaccine in China. Biologicals. 2006;34:265–272. doi: 10.1016/j.biologicals.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Morimoto K, Janssen WJ, Fessler MB, McPhillips KA, Borges VM, Bowler RP, Xiao YQ, Kench JA, Henson PM, Vandivier WR. Lovastatin enhances clearance of apoptotic cells (efferocytosis) with implications for chronic obstructive pulmonary disease. J Immunol. 2006;176:7657–7665. doi: 10.4049/jimmunol.176.12.7657. [DOI] [PubMed] [Google Scholar]
- 28.Moriya K, Todoroki T, Tsutsumi T, Fujie H, Shintani Y, Miyoshi H, Ishibashi K, Takayama T, Makuuchi M, Watanabe K, Miyamura T, Kimura S, Koike K. Increase in the concentration of carbon 18 monounsaturated fatty acids in the liver with hepatitis C: analysis in transgenic mice and humans. Biochem Biophys Res Commun. 2001;281:1207–1212. doi: 10.1006/bbrc.2001.4523. [DOI] [PubMed] [Google Scholar]
- 29.Nandi P, Charpilienne A, Cohen J. Interaction of rotavirus particles with liposomes. J Virol. 1992;66:3363–3367. doi: 10.1128/jvi.66.6.3363-3367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padilla-Noriega L, Werner-Eckert R, Mackow ER, Gorziglia M, Larralde G, Taniguchi K, Greenberg HB. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J Clin Microbiol. 1993;31:622–628. doi: 10.1128/jcm.31.3.622-628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. The global burden of diarrhoeal disease in children. Bull World Health Organ. 2003;81:236. [PMC free article] [PubMed] [Google Scholar]
- 32.Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;2:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapin C, Colard O, Delmas O, Tessier C, Breton M, Enouf V, Chwetzoff S, Ouanich J, Cohen J, Wolf C, Trugnan G. Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J Virol. 2002;76:4591–4602. doi: 10.1128/JVI.76.9.4591-4602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger MJ, Malfer C. Cerulenin blocks fatty acid acylation of glycoproteins and inhibits vesicular stomatitis and Sindbis virus particle formation. J Biol Chem. 1982;257:9887–9890. [PubMed] [Google Scholar]
- 35.Thorp EB, Gallagher TM. Requirements for CEACAMs and cholesterol during murine coronavirus cell entry. J Virol. 2004;78:2682–2692. doi: 10.1128/JVI.78.6.2682-2692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


