Abstract
Trionyx sinensis hemorrhagic syndrome virus (TSHSV) is a newly discovered lethal arterivirus that causes serious disease in Trionyx sinensis in China. In this study, the complete genome sequence of TSHSV was determined by RACE cloning, and the functions of the predicted proteins were predicted. The complete genome of TSHSV was found to be 17,875 bp in length, and a 3’-end poly(A) tail was detected. Eight TSHSV hypothetical proteins (TSHSV-HPs) were predicted by gene model identification. TSHSV-HP2, 3 and 4 were associated with replicase activity, since papain-like protease (PLPs), serine-type endopeptidase, P-loop-containing nucleoside triphosphate hydrolase, and EndoU-like endoribonuclease motifs were detected. Phylogenetic analysis showed that TSHSV clusters with an arterivirus from a Chinese broad-headed pond turtle.
Introduction
Trionyx sinensis hemorrhagic syndrome virus (TSHSV) was first discovered and isolated by Liu in our laboratory in 2013. This virus has recently become an important factor limiting the sustainable development of T. sinensis farming [5, 13]. When compared with previously discovered T. sinensis viruses, TSHSV appears to be more virulent, causing high mortality both in the wild and in captivity [5]. TSHSV-infected turtles display typical characteristics, namely breathing difficulty, interstitial pneumonia, and hyperemic laryngeal mucosa in the throat [13, 14]. An artificial infection experiment has demonstrated that TSHSV-infected turtles are seriously hemorrhagic in multiple organs, including liver, kidney and intestine [13]. These clinicopathological features induced by T. sinensis are consistent with those typically caused by arteriviruses, including porcine reproductive and respiratory syndrome virus (PRRSV), equine arteritis virus (EAV), and simian hemorrhagic fever virus (SHFV) [1, 4, 7, 11, 18]. In addition, Liu et al. cloned a 435-bp-length amplicon that showed sequence similarity at the amino acid level to the arteriviruses mentioned above [13]. Considering the extremely high degree of infectivity and pathogenicity of TSHSV, it is essential to do more research on this newly discovered virus. In this study, we cloned and analyzed the complete genome of TSHSV, and the biological functions of the encoded hypothetical proteins were predicted.
Complete genome sequencing and classification of TSHSV
T. sinensis turtles that began to show signs of disease in early September 2013 were collected from outdoor ponds of turtle farms in Zhejiang Province [15]. Total RNA was extracted from lungs using TRIzol Reagent (Invitrogen) following the manufacturer’s protocol. Integrated RNA with an OD260-to-OD280 ratio between 1.8 and 2.2 was used for subsequent cDNA synthesis. One µg of total RNA was reverse transcribed into 5’ and 3’ cDNA using a SMARTer® RACE 5’/3’ Kit (cat. no. 634858, Clontech, USA). To obtain the complete genome sequence of TSHSV, TSHSV-specific primers for RACE, namely TSHSV-5’ (5’-GATTACGCCAAGCTTGCTCCCTCTCAACAACCAGCCAAAC-3’) and TSHSV-3’ (5’-GATTACGCCAAGCTTTTCAGCCACTTGAGCCTGGTCCTTT-3’), were designed based on the previously reported partial sequence of TSHSV (GenBank accession no. MH447986). Each 50.0-μL reaction mixture contained 2.5 μL of 5’- or 3’-RACE-Ready cDNA, 15.5 μL of PCR-grade water, 25.0 μL of 2× SeqAmp buffer, 1.0 μL of SeqAmp DNA polymerase, 5.0 μL of 10× universal primer mix (UPM), and 1.0 μL of TSHSV-specific primer. The amplification conditions were as follows: 94 °C for 5 min, followed by 25 cycles of denaturation (94°C for 30 s), annealing (68°C for 30 s) and extension (72°C for 8 min). The PCR product was purified according to the operating manual using a Cycle Pure Kit (Omega) and send to Sangon Biotech Co. (Shanghai, China) for sequencing. The complete genome sequence of TSHSV was deposited in the GenBank database under accession number MH447987. The genome of TSHSV was found to be a positive-sense, single-stranded RNA with a 3’-end poly(A) tail. The whole TSHSV genome was 17,875 bp in length with a 1323-bp-length 5’-untranslated region (UTR) and an 828-bp-length 3’-UTR. Gene model identification (http://topaz.gatech.edu/GeneMark/) predicted that the TSHSV genome contained eight open reading frames (ORFs) encoding eight hypothetical proteins (HPs) (Table 1). These HPs were calculated to have lengths of 79, 308, 1552, 1692, 169, 194, 329 and 714 amino acids (aa), respectively.
Table 1.
Gene sequence characteristics and predicted functions of TSHSV proteins
| Protein name | Sequence information | Family membership | Homologous superfamilies | Molecular function | 3D template and description | ||||
|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | MW (kDa) | pI | Template | Similarity | Description | ||||
| TSHSV-HP1 | 237 | 8.70 | 4.86 | None predicted | None predicted | None predicted | 4hkr.1.A | 34% | Calcium-release-activated calcium channel protein 1 |
| TSHSV-HP2 | 924 | 35.4 | 8.65 | None predicted | None predicted | None predicted. | 4ium.1.A | 31% | papain-like protease 2 |
| TSHSV-HP3 | 4656 | 169.6 | 6.44 | Equine arteritis virus peptidase S32 (IPR008760, 887-1139) | None predicted |
serine-type endopeptidase activity (GO:0004252); 1a replicase protein (GO:0003824) |
5y4l.1.A | 32% | Non-structural protein |
| TSHSV-HP4 | 5076 | 188.0 | 7.02 | None predicted |
P-loop containing nucleoside triphosphate hydrolase (IPR027417, 798-1042); endoribonuclease EndoU-like (IPR037227, 1412-1528) |
zinc ion binding (GO:0008270) | 4n0n.1.A | 32% | Replicase polyprotein 1ab |
| TSHSV-HP5 | 507 | 18.7 | 10.75 | None predicted | None predicted | None predicted | 3d1d.1.A | 33% | RNA-induced transcriptional silencing complex protein tas3 |
| TSHSV-HP6 | 582 | 22.5 | 9.27 | None predicted | None predicted | None predicted | 4za1.1.A | 28% | NosA |
| TSHSV-HP7 | 987 | 37.1 | 7.59 | None predicted | None predicted | None predicted | 4p85.1.A | 29% | Est-Y29 |
| TSHSV-HP8 | 2142 | 80.4 | 7.30 | None predicted | None predicted | None predicted | 3sfm.1.A | 40% | Dihydrofolapfte reductase type 2 |
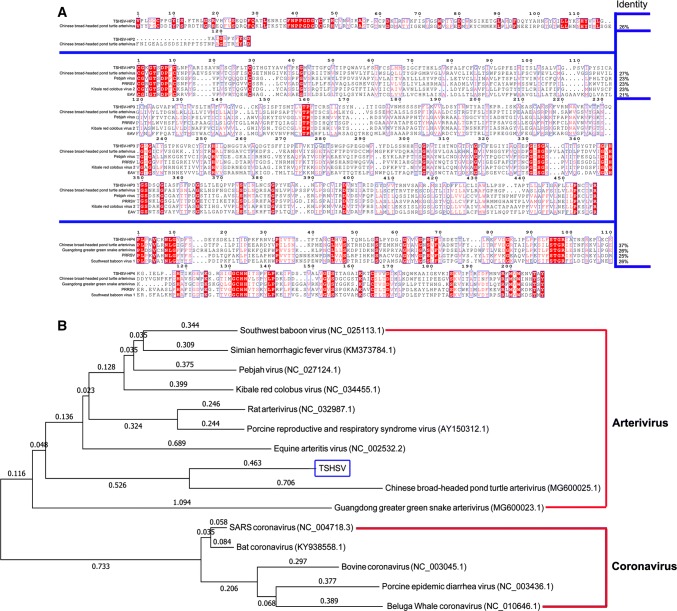
Conjoint analysis of a multiple amino acid sequence alignment and NCBI BLAST revealed that TSHSV-HP2, TSHSV-HP3 and TSHSV-HP4 shared partial sequence identity with some arteriviruses, including Chinese broad-headed pond turtle arterivirus, Pebjah virus, PRRSV, Guangdong greater green snake arterivirus, Wuhan Japanese halfbeak arterivirus and EAV [10]. The deduced amino acid sequences were most similar to those of Chinese broad-headed pond turtle arterivirus, while only 26%, 27% and 37% identity was calculated for HP2, HP3 and HP4, respectively (Fig. 1A). Phylogenetic analysis of the complete genome sequence of TSHSV revealed that TSHSV and Chinese broad-headed pond turtle arterivirus clustered in the same branch of the phylogenetic tree (Fig. 1B). In agreement with a previous analysis [13], the distinct genome and deduced amino acid sequences demonstrated that the TSHSV is a new arterivirus.
Fig. 1.
Multiple sequence alignment and phylogenetic analysis. A. Multiple amino acid sequence alignment of TSHSV-HP2, HP3 and HP4 with specific proteins of other viruses. The aligned sequences were as follows: 1ab protein (Chinese broad-headed pond turtle arterivirus; AVM87331.1); putative 1b protein (Pebjah virus; AKI29956.1); ORF1a polyprotein (porcine reproductive and respiratory syndrome virus; ADX07034.1); polyprotein (Kibale red colobus virus 2; AHH53949.1); ORF1ab polyprotein (equine arteritis virus; AAR14192.1); 1ab protein (Guangdong greater green snake arterivirus; AVM87321.1); ORF1b polyprotein, partial (porcine reproductive and respiratory syndrome virus; BAP16275.1); putative 1b protein (southwest baboon virus 1; YP_009067063.1). Identical (red) and similar (box) residues are indicated. B. Phylogenetic relationships between TSHSV, arteriviruses and coronaviruses based on the complete genome sequence. The tree was generated using the neighbor-joining method
Functional annotation of predicted proteins
The predicted protein sequences were analyzed using SwissProt (http://uniprot.org), GO (http://www.geneontology.org/) and three-dimensional structure analysis (http://www.expasy.ch/swissmod/SWISS-MODEL.html) to do functional annotation. TSHSV-HP2, TSHSV-HP3 and TSHSV-HP4 were found to contain domains involved in specific biological processes (Table 1). No specific functional domains were identified in the last five HPs. TSHSV-HP2 shared 31% sequence identity with papain-like protease (PLP) 2 at the amino acid sequence level. PLPs of coronaviruses are involved in processing virus-encoded large replicase polyproteins and also function as deubiquitinating enzymes [19]. This protease has been shown to be synthesized during infection with many coronaviruses, including human coronavirus NL63 (HCoV-NL63) [6], murine hepatitis virus-A59 (MHV-A59) [3], and severe acute respiratory syndrome coronavirus (SARS-CoV) [2], and these viruses are capable of synthesizing different amount of PLPs. The genome of the representative arterivirus PRRSV also contains a region encoding a papain-like cysteine protease alpha (PCPalpha) domain, which gives further support for the potential classification of TSHSV as an arterivirus [17].
TSHSV-HP3 appears to belong to the same protein family as EAV peptidase S32 (IPR008760). Additional molecular features of proteins with serine-type endopeptidase activity (GO: 0004252) and 1a replicase protein activity (GO: 0003824) were also found in this protein. Serine-type endopeptidases are often involved in viral protein processing. Proteolytic enzymes that require a serine for their catalytic activity are ubiquitous and can be found in viruses, bacteria and eukaryotes [9]. They can be associated with a wide range of peptidase activities including exopeptidase, endopeptidase, oligopeptidase and omega-peptidase activity [8]. For the arteriviruses, EAV, PRRSV-I, PRRSV-II, lactate dehydrogenase-elevating virus (LDV) and SHFV, a serine protease domain is present in non-structural protein 4 (nsp4), a 21-kDa cleavage product from the central region of the ORF1a-encoded polypeptide [15, 16]. It has been demonstrated that this is the main proteinase responsible for producing the majority of non-structural proteins (nsps) from the polyproteins (pp1a and pp1ab). Furthermore, it is responsible for the expression of replicase proteins, [12] suggesting that a similar function of TSHSV-HP3.
TSHSV-HP4 is predicted to have replicase activity, since a P-loop containing a nucleoside triphosphate hydrolase domain (IPR027417) and an EndoU-like endoribonuclease domain (IPR037227) were detected at positions 612-689 and 798-1042 respectively, of this protein (Table 1). These two functional activities have been confirmed for the replicase polyprotein 1ab of PRRSV [20].
In summary, as the full-length genome of a new arterivirus, TSHSV, was cloned and verified to be a positive-sense, single-stranded RNA with a poly(A) tail at its 3’ end. The encoded proteins TSHSV-HP2, TSHSV-HP3 and TSHSV-HP4 were predicted to be involved in replicase activity, whereas the functions of the last five HPs remain unknown. Further functional analysis and validation tests need to be conducted to determine the functions of these proteins.
Acknowledgements
We would like to thank the Huzhou Fisheries Technical Extension Station for providing ill T. sinensis.
Funding
This study was partially funded by the Zhejiang Provincial Science and Technology Program (Project no. 2018F10013) and the Zhejiang Provincial Natural Science Foundation (Project no. LY15C190001).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed by the authors. Samples were obtained from a T. sinensis farm, and this study did not include animal experiments.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Balasuriya UBR, Go YY, MacLachlan NJ. Equine arteritis virus. Vet Microbiol. 2013;167:93–122. doi: 10.1016/j.vetmic.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonilla PJ, Hughes SA, Piñón JD, Weiss SR. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 4.Bøtner A, Nielsen J, Bille-Hansen V. Isolation of porcine reproductive and respiratory syndrome (PRRS) virus in a Danish swine herd and experimental infection of pregnant gilts with the virus. Vet Microbiol. 1994;40:351–360. doi: 10.1016/0378-1135(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Li D, Fang J, Zhang L, Gu Y, Liu L, Zhang G. Apolipoprotein mediates soft-shelled turtle systemic septicemia spherical virus (STSSSV) infection. Aquaculture. 2017;473:501–507. doi: 10.1016/j.aquaculture.2017.03.022. [DOI] [Google Scholar]
- 6.Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coignoul FL, Cheville NF. Pathology of maternal genital tract, placenta, and fetus in equine viral arteritis. Vet Pathol. 1984;21:333–340. doi: 10.1177/030098588402100311. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande MRATMGV. Molecular and biotechnological aspects of microbial proteases. FEMS Microbiol Rev. 2010;23:411–456. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhillon A, Sharma K, Rajulapati V, Goyal A. Proteolytic enzymes. Amsterdam: Elsevier; 2017. [Google Scholar]
- 10.EJST, Yang H, Yoo D (2018) Family Arteriviridae. Virus Genes
- 11.Johnson RF, Dodd LE, Yellayi S, Gu W, Cann JA, Jett C, Bernbaum JG, Ragland DR, St CM, Byrum R. Simian hemorrhagic fever virus infection of rhesus macaques as a model of viral hemorrhagic fever: clinical characterization and risk factors for severe disease. Virology. 2011;421:129. doi: 10.1016/j.virol.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dinten LC, Gorbalenya AE, Snijder EJ. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Cao Z, Lin F, Ye X, Xu Y. Partial sequence of a novel virus isolated from Pelodiscus sinensis hemorrhagic disease. Intervirology. 2015;58:197–204. doi: 10.1159/000437354. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Cao Z, Lin F, Ye X, Lu S, Lyv S. The histopathological characteristics caused by Trionyx sinensis Hemorrhagic syndrome virus (TSHSV) and comparative proteomic analysis of liver tissue in TSHSV-Infected Chinese Soft-Shelled turtles (Pelodiscus sinensis) Intervirology. 2017;60:19–27. doi: 10.1159/000479795. [DOI] [PubMed] [Google Scholar]
- 15.MacLachlan NJ, Dubovi EJ. Fenner’s veterinary virology. 5. Amsterdam: Elsevier; 2017. [Google Scholar]
- 16.Snijder EJ, Gorbalenya AE. Handbook of proteolytic enzymes. 3. Amsterdam: Elsevier; 2013. [Google Scholar]
- 17.Sun Y, Xue F, Guo Y, Ma M, Hao N, Zhang XC, Lou Z, Li X, Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J Virol. 2009;83:10931. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu-Bao LI, Meng XL, Fei MA (2009) Distribution in tissues and pathological lesions of PRRSV to piglets. Heilongjiang Anim Sci Vet Med :19–21
- 19.Yu L, Zhang X, Wu T, Wang Y, Meng J, Liu Q, Niu X, Wu Y. The papain-like protease of avian infectious bronchitis virus has deubiquitinating activity. Arch Virol. 2017;162:1–8. doi: 10.1007/s00705-016-3065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang MFWW, Chen ZZ (2017) Structure and Function of PRRSV nsp11 Endoribonuclease. In: 2017 International Conference on Medicine Sciences and Bioengineering, pp 89–94