Abstract
The Eurasian otter (Lutra lutra Linnaeus, 1758), a predator from the Order Carnivora, Family Mustelidae, evolved the ability to swim and forage in water, being an important element of biodiversity. Otters are widely spread through Portugal, and scats have been extensively used in ecology studies; however, valid information on their microbiota is scarce. This work represents a first approach to characterise the otter faecal microflora in samples collected in river stretches of the Sado river basin (Portugal) during winter 2006. Eight sampling stretches of 8 km were selected, and from each, six to eight sampling sites were visited. A total of 31 scats were analysed. The microflora studied included aerobic bacteria, spore-forming anaerobic bacteria and viruses (coronavirus, parvovirus, adenovirus, parainfluenza virus). Bacterial isolates were identified based on morphology and metabolic pathways, and virus detection was performed by polymerase chain reaction. The results revealed the high degree of bacterial diversity in the faecal microflora of L. lutra. A total of 88 Gram-negative (23 genera) and 44 Gram-positive isolates (ten genera) were identified. The identification of four isolates was inconclusive, and their identification was performed by 16S ribosomal ribonucleic acid sequencing, which confirms the need for biochemical testing optimisation regarding animal isolates. None of the scats was positive for virus detection. Identification of otter faecal microflora and of potential pathogens is an important first step towards understanding and monitoring their importance in otter population health.
Keywords: Bacteria, Identification, Otter, Virus
Introduction
The otter (Lutra lutra Linnaeus, 1758) is one of the few Eurasian predators (Carnivora, Mustelidae) that evolved the ability to actively swim and forage in water, rendering it an important element of biodiversity. Otters are well spread through Portugal (Trindade et al. 1998) and may be found in a wide range of aquatic environments, from very small streams and large rivers to dams, including also coastal and estuarine environments (Santos-Reis et al. 1995). Otter densities in Portugal are unknown but suspected to be high (Santos-Reis et al. 2003), in opposition to many European countries, where otter populations progressively declined since the beginning of the twentieth century (Mason and Macdonald 1986) and only now are apparently recovering (Cortés et al. 1998).
The need for genetic, demographic and life-history information about this species is thus a reality (Kohn and Wayne 1997; Kruuk 2006). Individual molecular identification through faecal deoxyribonucleic acid (DNA) analysis is a new non-invasive method being applied in abundance and home-range studies (Dallas et al. 2003; Arrendal et al. 2004; Selkoe and Toonen 2006). To improve the success of DNA extraction from the faeces, scats need to be collected fresh, and to ensure this, surveys are usually repeated in consecutive days at sunrise. Fresh samples also allow the characterisation of the faecal microflora (Kohn and Wayne 1997).
Valid microflora characterisation studies on otters are scarce, and little is known about the clinical significance of faecal isolates and environmental contaminant-related diseases. Otters can be affected by viral, bacterial, mycotic and parasitic diseases. Most studies regarding bacterial diseases in otters concern the American river otters (Lutra canadensis). These are shown to be sensitive to genitourinary infections promoted by Proteus mirabilis, beta-haemolytic Streptococcus and beta-haemolytic Escherichia coli (Hoover and Tyler 1986). The presence of Klebsiella pneumoniae has been related to retropharyngeal abscesses (Hoover and Tyler, 1986). Regarding the Eurasian otters, they were shown to be a possible natural reservoir for pathogenic Yersinia enterocolitica and Y. pseudotubeculosis, playing an important role in the epidemiology of yersinioses (Nikolova et al. 2001). Studies regarding southern sea otters (Enhydra lutris) infectious diseases and necropsy data are also available (Smith et al. 2002; Hanni et al. 2003; Kreuder et al. 2003).
Otters may be susceptible to several canine and feline viral pathogens. Although the presence of antibodies against canine and feline parvovirus, canine and feline coronavirus, canine herpesvirus and canine adenovirus (CAD) type 2 have been reported (Kimber and Kollias 2000; Kimber et al. 2000), the potential role of these species as reservoirs of infectious virus has not been investigated.
In Portugal, the otter health status is unknown. This work represents a first approach to the study of viable otter faecal microflora in several river stretches of the Sado river basin (Portugal). Bacteria were identified from stool swabs on the basis of morphology and metabolic pathways or by partial 16S ribosomal DNA (rDNA) sequencing. To collect information about the presence and prevalence of canine viruses in the population, a virological survey was also conducted for Canine Parvovirus (CPV; Parvoviridae), Canine Parainfluenza Virus (CPIV; Paramyxoviridae) and CAD1 and CAD2 (Adenoviridae) on stool samples.
Materials and methods
Scat sampling
The Sado river basin is located in Alentejo province, in the south of Portugal (Fig. 1). Eight river stretches, with approximately 8 km each and spread throughout the basin to represent the several types of existing streams (ditches to main river) were chosen for sampling. Chosen river stretches were sufficiently far apart to ensure use by different otters. At each stretch, six to eight otter marking places were chosen for scat collection. In winter of 2006, these sampling sites were surveyed at sunrise, for 5 consecutive days, assuring that sample collection was performed within 15 h from deposition by the otter. Otter scats are easily recognisable to the trained eye and are usually deposited by the otter in prominent places as marking activity for territory defence. Scats collected at the river stretches might be from the same individual, but scats from different river stretches are probably not. Among 119 scats collected, 31 were randomly selected on site and processed for microflora analysis (Table 1), being the remaining used in diet assessment and in the molecular identification of individual otters (data not shown). For each scat sample selected, an AMIES swab (FL medical) was immediately performed for bacteria identification and kept refrigerated until further processing. The remaining scat sample was kept frozen for viral detection.
Fig. 1.
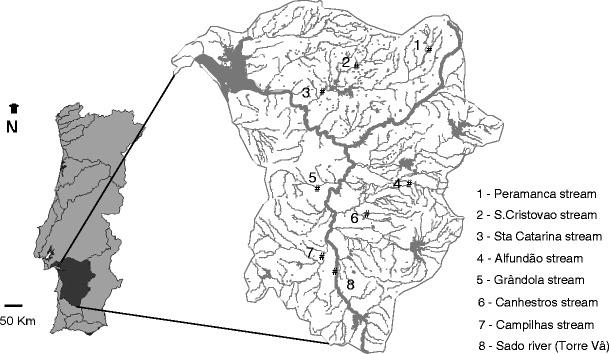
Sado river basin (in dark grey) and location of surveyed streams
Table 1.
Surveyed streams in river Sado basin and faecal otter samples collected at each stream
| Stream | Samples |
|---|---|
| “Ribeira de Sta Catarina” | L1, L2, L3, L5, L8 |
| “Ribeira de S.Cristóvão” | L4, L6, L7 |
| “Ribeira de Peramanca” | L9, L10 |
| “Rio Sado” | L11, L12, L13, L14, L15 |
| “Ribeira de Alfundão” | L16 L17, L18, L24 |
| “Ribeira de Canhestros” | L19, L20, L21, L22, L23 |
| “Ribeira de Campilhas” | L25, L26, L27, L28, L29 |
| “Ribeira de Grândola” | L30, L31 |
Bacteria detection and phenotypic identification
The bacteria studied included aerobic bacteria and anaerobic spore-forming bacteria. Aerobic bacteria isolation from swabs was performed in Columbia agar (bioMérieux), incubated at 37°C for 24 to 48 h. The swabs were also inoculated in Brain Heart Infusion broth (Merck), subjected to heat shock (80°C, 10 min), inoculated in Schaedler agar (bioMérieux) and incubated for 48 h at 37°C in the absence of oxygen for spore-forming anaerobic bacteria detection. Isolates were identified through their macro- and microscopic morphology, staining characteristics and biochemical profiles using API (bioMérieux) and BBL Crystal (Becton Dickinson) galleries, according to the manufacturer’s instructions.
Bacterial isolates which biochemical profiles that originated doubtful identifications were identified through 16S rDNA based analysis. Bacterial cultures were grown in Nutrient Broth (Difco) supplemented with 1.5% agar for 24–36 h at 37°C. A colony was collected with a sterile toothpick and suspend in 100 μL of a lysis buffer containing 10 mM Tris, 1 mM ethylenediamine tetraacetic acid, 0.1% Tween 20, pH 8.0. Cell lysates were prepared by heating at 100°C for 10 min followed by rapid cooling on ice. An 800-bp segment of the 16S ribosomal ribonucleic acid (RNA) gene was amplified using the universal primers 104F and 907R (Muyzer et al. 1996). Polymerase chain reaction (PCR) amplification was performed in a T gradient thermocycler (Biometra) as described by Chambel et al. (2006). The amplification products were purified with a JETquick spin column (Genomed) and sequenced in a CEQ 2000-XL automated DNA capillary sequencer (Beckman Coulter) by a dye-labelled dideoxy-termination method using the 907R primer with standard protocols (Dye Terminator Cycle Sequencer start kit, Beckman Coulter; Chambel et al. 2006). Partial 16S rDNA sequences with approximately 600 bp were compared with National Center for Biotechnology Information/GenBank entries using the Basic Local Alignment Search Tool algorithm (Altschul et al. 1997). Species level identification was assumed for homology values higher than 97%.
Virus detection and PCR identification
Viral DNA and RNA were extracted simultaneously from 31 stool samples with QIAamp MinElute Virus Spin Kit (Qiagen) according to the manufacturer’s instructions. The positive controls for CPIV RNA, CAD2 DNA and CPV DNA were extracted from Vanguard 7 vaccine (Pfizer) with QIAamp MinElute Virus Spin Kit (Qiagen). The lyophilised fraction of the vaccine was ressuspended in 1 mL of the liquid fraction, and 100 μL was used for viral nucleic acid extraction, corresponding to 106 tissue culture 50% infectious dose (TCID50) CPIV strain NL-CPI5, 103.2 TCID50 CAD2 strain Manhattan and 107 TCID50 CPV strain NL35D.
For reverse transcription (RT) PCR and PCR assays, the primers sequence and the expected size of the amplicons are indicated in Table 2. For amplification of CPIV RNA, one-step RT-PCR reactions were performed according to Desario et al. 2005, in 50 μL with 10 μL of the sample and 50 pmol of each primer using AccessQuick™ RT-PCR System (Promega) according to manufacturer’s instructions. The nested PCR was done in 50 μL using FideliTaq™ PCR Master Mix (2×) (USB) according to the manufacturer’s instructions, 2 μL of the RT-PCR reaction and 50 pmol of each primer using the same PCR cycling conditions (Desario et al. 2005). DNA amplification of CAD1 and CAD2 was done in 50 μL using FideliTaq™ PCR Master Mix (2×) (USB), 10 μL of sample and 50 pmol of each primer, according to Desario et al. 2005. For DNA amplification of CPV, the reaction was done in 50 μL using FideliTaq™ PCR Master Mix (2×) (USB), 2 mM MgCl2 final volume, 10 μL of sample and 50 pmol of each primer, according to Erles et al. 2004.
Table 2.
| Virus/primers | Primer sequence (5′–3′) | Nucleotide position | Amplicon size |
|---|---|---|---|
| Canine Parainfluenza virus (CPIV) | |||
| CPI1 | AGTTTGGGCAATTTTTCGTCC | 120–140a | 667 bp |
| CPI2 | TGCAGGAGATATCTCGGGTTG | 786–766a | |
| CPI3 | CGTGGAGAGATCAATGCCTATGC | 521–543a | 187 bp |
| CPI4 | GCAGTCATGCACTTGCAAGTCACTA | 702–678a | |
| Canine Adenovirus 1 (CAV1) and 2 (CAV2) | |||
| CA 1,2 F | CTGGGCGGGATTTAGAGGGTGG | 18,804–18,825b | 704 bp |
| CA 1,2 R | CAAGGGCGTGGGCGGAGTTAGA | 19,507–19,486b | |
| Canine Parvovírus (CPV) | |||
| CP1 | CAGGAAGATATCCAGAAGGA | 4,003–4,022c | 583 bp |
| CP2 | GGTGCTAGTTGATATGTAATAAACA | 4,561–4,585c | |
Serial dilutions were performed using the positive control of CPIV RNA, CAD1, CAD2 and CPV DNA, to determine the sensitivity of the RT-PCR and the PCR assays for detection of viral nucleic acid in the biological materials. The viral nucleic acids were diluted until an end point dilution corresponding to 1 TCID50 of each and subjected to RT-PCR and PCR.
Results
A total of 132 bacterial isolates were obtained. Forty-four (33.3%) were Gram-positive bacteria, belonging to ten genera, and 12 isolates corresponded to spore-forming anaerobic bacteria (9.1%). More than one species was isolated for four genera (Table 3). It was also possible to detect 88 Gram-negative isolates (66.7%), corresponding to 23 genera, and for ten genera, more than one species were isolated (Table 4).
Table 3.
Gram-positive bacteria isolated from otter scat samples (river Sado basin)
| Genus | Species | Number of isolates | Number of positive samples |
|---|---|---|---|
| Aerococcus | A. viridans | 2 (1.5%) | 2 |
| Bacillus | B. brevis, B. cereus, B. licheniformis, B. megaterium, B. pumilus, B. sphaericus, Bacillus sp. | 12 (9.1%) | 8 |
| Cellulomonas/ Microbacterium | Cellulomonas/Microbacterium sp. | 2 (1.5%) | 2 |
| Clostridium a, b, c | C. beijerinckii/butyricum, C. perfringens, C. sordelii, Clostridium sp. | 12 (9.1%) | 10 |
| Corynebacterium a, d, e | C. aquaticum, C. diphteriae mitis | 3 (2.3%) | 2 |
| Enterococcus | E. durans, E. faecalis, E. faecium | 5 (3.8%) | 5 |
| Paenibacillus | P. alvei | 1 (0.8%) | 1 |
| Propionibacterium | P. avidum | 3 (2.3%) | 3 |
| Staphylococcus a, f | S. simulans | 1 (0.8%) | 2 |
| Streptococcus a, b, c, g | S. porcinus | 1 (0.8%) | 1 |
| No identification | 2 (1.5%) | 2 | |
| Total | 44 (33.3%) |
Table 4.
Gram-negative bacteria isolated from otter scat samples (river Sado basin)
| Genus | Species | Number of isolates | Number of positive samples |
|---|---|---|---|
| Acinetobacter a, b, c | A. lwoffi | 1 (0.8%) | 1 |
| Aeromonas a, b | A. hydrophila, A. hydrophila/caviae, A. sobria | 18 (13.6%) | 15 |
| Agrobacterium | A. tumefaciens | 1 (0.8%) | 1 |
| Burkholderia | B. cepacia | 4 (3.0%) | 4 |
| Buttiauxella | B. agrestis | 1 (0.8%) | 1 |
| Chryseomonas | C. luteola | 1 (0.8%) | 1 |
| Citrobacter | C. amalonaticus, C. braaki, C. youngae, Citrobacter sp. | 7 (5.3%) | 6 |
| Empedobacter | E. brevis | 1 (0.8%) | 1 |
| Enterobacter | E. amnigenus, E. cancerogenus, E. cloacae, E. gergoviae, E. sakazakii, Enterobacter sp. | 7 (5.3%) | 7 |
| Escherichia a, d | E. coli, E. vulneris | 3 (2.3%) | 3 |
| Hafnia | H. alvei | 2 (1.5%) | 2 |
| Klebsiella a, d | K. oxytoca, K. pneumoniae, K. ozaenae | 3 (2.3%) | 3 |
| Moraxella | Moraxella sp. | 2 (1.5%) | 2 |
| Morganella | M. morganii | 1 (0.8%) | 1 |
| Pantoea | P. agglomerans, Pantoea spp. | 13 (9.8%) | 12 |
| Pasteurella a, e | Pasteurella sp. | 1 (0.8%) | 1 |
| Pseudomonas a, b, c | P. aeruginosa, P. fluorescens, P. putida | 5 (3.8%) | 5 |
| Rahnella | R. aquatilis | 1 (0.8%) | 1 |
| Salmonella a, c, f | S. arizonae, S. pullorum | 4 (3.0%) | 3 |
| Serratia | S. fonticola, S. plymuthica, Serratia sp. | 3 (2.3%) | 2 |
| Shigella | Shigella sp. | 1 (0.8%) | 1 |
| Vibrio | V. alginolyticus, V. metschikovii, V. parahaemolyticus | 5 (3.8%) | 5 |
| Yokenella | Y. regensburgei | 1 (0.8%) | 1 |
| No identification | 2 (1.5%) | 2 | |
| Total | 88 (66.7%) |
It was not possible to identify four isolates (3.0%) based on their phenotypic characteristics, and their identification was performed by 16S rDNA sequencing (Table 4). The Gram-positive isolates were identified as Exiguobacterium sp., and the Gram-negative isolates were identified as Y. enterocolitica and Citrobacter sp.
The biochemical profiles of the E. coli isolates also originated doubtful identifications. Although isolate L15-5 was identified as E. coli with a significance of 99.3%, the profile (5045552) originated a doubtful identification. In relation to isolate L18-4, a BBL E/NF system (5465646171, 89.24%) was performed, as the API profile (5446572) was unacceptable. Therefore, these two isolates were subjected to 16S rDNA sequencing (Table 5). This method revealed that the isolates could be identified as Shigella boydii (99%), E. coli (99%) or Photorhabdus luminescens (99%).
Table 5.
Species level identification by partial 16S rDNA sequencing
| Isolate identification | Partial 16S rDNA (bp) | BLAST results | ||
|---|---|---|---|---|
| Accession number | Taxonomic description | Percent homology | ||
| L2-5 | 619 | GI89513049 | Exiguobacterium sp. | 100 |
| L5-5 | 620 | GI71725879 | Exiguobacterium acetylicum | 99 |
| GI34536531 | Exiguobacterium oxidotolerans | 99 | ||
| L6-3 | 575 | GI861167 | Yersinia enterocolitica | 100 |
| GI46406312 | Yersinia frederiksenii | 99 | ||
| GI13991902 | Yersinia rohdei | 98 | ||
| L7-4 | 619 | GI3169777 | Citrobacter sp. | 99 |
| GI90655953 | Buttiauxella agrestis | 98 | ||
| GI70610311 | Pantoea sp. | 98 | ||
| L15-5 | 617 | GI52697039 | Shigella boydii | 99 |
| L18-4 | 618 | GI46242278 | Escherichia coli | 99 |
| GI38112705 | Photorhabdus luminescens | 99 | ||
Regarding virus detection, the sensitivity of the RT-PCR and the PCR assays was determined by the amplification of serial dilutions of the positive control of CPIV RNA, CAD1, CAD2 and CPV DNA. All the dilutions were amplified showing high specificity and sensitivity. The 31 samples were tested for the three viral pathogens separately. None was positive showing the absence of these viral pathogens in the analysed biological material.
Discussion
In recent years, there has been a growing interest in wildlife diseases because of public concern for its welfare, human medical interest in zoonoses, their potential role as environmental pollution monitors and veterinary interest in wildlife and their potential role as reservoirs of infection for farm or companion animals (Simpson 2000). We aimed at investigating microbial diversity of the otter’s faecal flora, as they are well spread in this country, being found in a wide range of aquatic environments. Although cultured-based methods applied for faecal bacteriological examination have several limitations (Leser et al. 2002), as there are no data available on the microbiota of the Portuguese otter, we have chosen not to apply selective media or to perform PCR-based analysis, which would restrict microbiota characterisation. Harmsen et al. (1999) have already stated that molecular biology methods are sometimes insufficient for characterisation of faecal bacteria and provide no information on viability of microorganisms. Nevertheless, it should be noticed that fastidious bacteria, like Mycobacterium sp., would be lost in our study and acid-fast staining does not provide accurate results. Further studies should address this pathogen, by PCR amplification.
In the present study, it was possible to identify a total of 132 isolates. One third of the isolates (33.3%) corresponded to Gram-positive bacteria, and of these, 17 have been considered as potentially pathogenic: Clostridium (9.1%), Corynebacterium (2.3%), Staphylococcus (0.8%) and Streptococcus (0.8%). References in the literature regarding the potential pathogenic role of Corynebacterium spp. in otters could not be found. The pathogenic role of haemolytic staphylococci to European river otters was described by Foster et al. (1997), while Hoover and Tyler (1986) related the occurrence of genitourinary infections with beta-haemolytic Streptococcus. Staphylococcus were also related to hepatic haematoma, secondary infections from the lung, kidney, and spleen and subcutaneous abscesses by Kimber and Kollias (2000), who also described the presence of toxinogenic C. perfringens in the North American river otters.
Most of the isolates were Gram-negative bacteria (66.7%) that were found in all but two of the swab samples. Because these bacteria are sensitive to collection and transport conditions, this indicates that isolation success was not affected by sampling procedures (Oliveira et al. 2007). From the 23 genera isolated, seven corresponded to potential Gram-negative pathogenic isolates according to their biochemical profile: Acinetobacter lwoffi (0.8%), Aeromonas spp. (13.6%), Escherichia spp. (2.3%), Klebsiella spp. (2.3%), Pasteurella sp. (0.8%), Pseudomonas spp. (3.8%) and Salmonella spp. (3.0%).
References in the literature regarding the potential pathogenic role of Acinetobacter spp. and Pasteurella spp. in otters could not be found. Kimber and Kollias (2000) described the association of Acinetobacter with otters, but they could not determine its clinical significance. The pathogenic role of K. pneumoniae to otters was discussed by Hoover and Tyler (1986), who described these bacteria as responsible for otters’ mortality by retropharyngeal abscess. These authors also described the occurrence of genitourinary infection of female otters because of pathogenic E. coli. The potential pathogenic role of Pseudomonas spp. must be taken into account, although the only description of otter disease was associated with Pseudomonas putrefaciens (Kimber and Kollias 2000). Salmonella spp. were already isolated from otters at rehabilitation centres (Smith et al. 2002) and in the wild (Kimber and Kollias 2000), which suggests that wildlife may play a reservoir role for Salmonella infections.
Conclusions about the qualitative constitution of the otter faecal flora should take in consideration the otters’ diet, as nutrient metabolism influences the establishment and maintenance of mammalian intestine bacteria (Hooper et al. 2002). Otters diet was characterised by macroscopic observation of the scats (data not shown), being verified that the major constitutes of the otters diet vary between crayfish (61.3% of the scats), fish (29.0%) and fish and crayfish (9.7%). No association was found between diet and microflora. The distribution of the isolates should also be taken in consideration. Our study describes the cultivable aerobic and anaerobic spore-forming bacteria from 31 scats, and we do not believe that this sample size may be representative of the otter faecal flora, especially to differentiate between a fully established bacterial community and transient microorganisms. Data on more animals from different sampling sites and a more thorough analysis may allow for discriminating between faecal flora and transient bacteria.
In this work, we also confirmed the urgent need to optimise biochemical identification galleries regarding animal isolates, which is supported by the fact that it was not possible to identify four isolates based on their biochemical profiles and also by the difficulties observed in relation to the E. coli biochemical identification. One of these isolates was identified as belonging to the Yersinia genus, which has already been isolated from lung samples of Eurasian otters (Nikolova et al. 2001).
Viral diseases have been reported in river otters. The presence of antibodies against feline and canine virus was already referred by other authors (Kimber and Kollias 2000; Kimber et al. 2000) in river otters, indicating their susceptibility to viral infection. A vast number of pathogen sequences are available for amplification from faecal material (Kohn and Wayne 1997). None of the samples was positive for the viral nucleic acids tested in this work. CPIV, CAD1, CAD2 and CPV are eliminated in the stools and persist in the environment for long periods because of their resistance to adverse climatic conditions (Greene 1990). The negativity to all three viruses in the processed stool samples indicates that these animals were not excreting the virus at the time of collection. Infection by Parvovirus is more frequent in the beginning of spring and through out the summer (Greene 1990). The Adenovirus and Parainfluenza virus are ubiquitous, but their pathogenic impact is higher in populations with high animal density, which is not the case with the Eurasian otter, as they are solitary territorial animals. Further studies should investigate if this negativity is related to the time of collection, as the PCR inhibitors that are usually encountered in faeces (Kohn and Wayne 1997) were eliminated by the DNA extraction method. Amplification of short DNA segments (from 187 to 704 bp) also results in more consistent amplifications, eliminating the possibility of no amplification because of the fact that DNA in faeces is present in low copy numbers or degraded (Kohn and Wayne 1997). The success of DNA extraction was also improved by the collection of scats at sunrise, to assure their freshness.
Identification of pathogens responsible for substantial morbidity and mortality in otters and the geographic distribution of these pathogens is an important first step towards understanding and monitoring their importance in otter population health. Further studies should include fingerprinting of potentially pathogenic isolates, which may allow the identification of the contamination source, contributing to the development management plans to minimise environmental contaminations.
Acknowledgements
This study was conducted with the financial support of the “Centro de Investigação Interdisciplinar em Sanidade Animal” (CIISA/FMV) from Faculdade de Medicina Veterinária, (Faculty of Veterinary Medicine, Technical University of Lisbon) and of the European Union (contract number EVK2-CT-2002-00142-FRAP). M. Oliveira and T. Sales-Luís hold scholarships from “Fundação para a Ciência e Tecnologia” (SFRHBPD/23226/05 and SFRH/BD/5163/2001).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrendal J, Walker CW, Sundqvist AK, Hellborg L, Vilal C. Genetic evaluation of an otter translocation program. Conservation Genetics. 2004;5:79–88. doi: 10.1023/B:COGE.0000014059.49606.dd. [DOI] [Google Scholar]
- Chambel L, Chelo IM, Zé-Zé L, Pedro LG, Santos MA, Tenreiro R. Leuconostoc pseudoficulneum sp. nov., isolated from a ripe fig. Int J Syst Evol Microbiol. 2006;56:1375–1381. doi: 10.1099/ijs.0.64054-0. [DOI] [PubMed] [Google Scholar]
- Cortés Y, Fernández-Salvador R, Garcia F, Virgós E, Llorente M. Changes in otter Lutra lutra distribution in Central Spain in the 1964–1995 period. Biol Conserv. 1998;86:179–183. doi: 10.1016/S0006-3207(98)00009-3. [DOI] [Google Scholar]
- Coyle MB, Lipsky BA. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin Microbiol Ver. 1990;3:227–246. doi: 10.1128/cmr.3.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas JF, Coxon KE, Sykes T, Chanin PRF, Marshall FM, Carss DN, Bacon PJ, Piertney SB, Racey PA. Similar estimates of population genetic composition and sex ratio derived from carcasses and faeces of Eurasian otter Lutra lutra. Mol Ecol. 2003;12:275–282. doi: 10.1046/j.1365-294X.2003.01712.x. [DOI] [PubMed] [Google Scholar]
- Desario C, Decaro N, Campolo M, Cavalli A, Cirone F, Elia G, Martella V, Lorusso E, Camero M, Buonavoglia C. Canine parvovirus infection: which diagnostic test for virus? J Virol Methods. 2005;126:179–185. doi: 10.1016/j.jviromet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Erles K, Dubovi EJ, Brooks HW, Brownlie J. Longitudinal study of viruses associated with canine infectious respiratory disease. J Clin Microbiol. 2004;42:4524–4529. doi: 10.1128/JCM.42.10.4524-4529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster G, Ross HM, Hutson RA, Collins MD. Staphylococcus lutrae sp. nov., a new coagulase-positive species isolated from otters. Int J Syst Bacteriol. 1997;47:724–726. doi: 10.1099/00207713-47-3-724. [DOI] [PubMed] [Google Scholar]
- Funke G, Von Graenevitz A, Clarridge JE, Bernard KA. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC. Infectious diseases of the dog and cat. Philadelphia: Saunders; 1990. p. 934. [Google Scholar]
- Hanni KD, Mazet JAK, Gulland FMD, Estes J, Staedler M, Murray MJ, Miller M, Jessup DA. Clinical pathology and assessment of pathogen exposure in southern and Alaskan sea otter. J Widl Dis. 2003;39:837–850. doi: 10.7589/0090-3558-39.4.837. [DOI] [PubMed] [Google Scholar]
- Harmsen HJM, Gibson GR, Elfferich P, Raangs GC, Wildeboer-Veloo ACM, Argaiz A, Roberfroid MB, Welling GW. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol Lett. 1999;183:125–129. doi: 10.1111/j.1574-6968.2000.tb08945.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Hoover JP, Tyler RD. Renal function and fractional clearances of American river otters (Lutra canadensis) J Wildl Dis. 1986;22:547–556. doi: 10.7589/0090-3558-22.4.547. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Schulze-Tanzil G, Martel J-L, Chaslus-Dancla E, Schwarz S. Antimicrobial resistance in Pasteurella and Mannheimia: epidemiology and genetic basis. Vet Res. 2001;32:323–339. doi: 10.1051/vetres:2001128. [DOI] [PubMed] [Google Scholar]
- Kimber KR, Kollias GV. Infectious and parasitic diseases and contaminant-related problems of North American river otters (Lontra canadensis): a review. J Zoo Wildl Med. 2000;31:452–472. doi: 10.1638/1042-7260(2000)031[0452:IAPDAC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kimber KR, Kollias GV, Dubovi EJ. Serologic survey of selected viral agents in recently captured wild North American river otters (Lontra canadensis) J Zoo Wildl Med. 2000;31:168–175. doi: 10.1638/1042-7260(2000)031[0168:SSOSVA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Wayne RK. Facts from feces revisited. TREE. 1997;12:223–227. doi: 10.1016/s0169-5347(97)01050-1. [DOI] [PubMed] [Google Scholar]
- Kreuder C, Miller MA, Jessup DA, Lowenstine LJ, Harris MD, Ames JA, Carpenter TE, Conrad PA, Mazet JAK. Patterns of mortality in southern sea otters (Enhtdra lutris nereis) from 1998–2001. J Wild Dis. 2003;39:495–509. doi: 10.7589/0090-3558-39.3.495. [DOI] [PubMed] [Google Scholar]
- Kruuk H. Otters: ecology, behaviour and conservation. Oxford: Oxford University Press; 2006. p. 265. [Google Scholar]
- Leser TD Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. Culture-Independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Envir Microbiol. 2002;68:673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C, MacDonald S. Otters: ecology and conservation. Cambridge: Cambridge University Press; 1986. p. 236. [Google Scholar]
- Muyzer G, Hottentrager S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA – a new molecular approach to analyse the genetic diversity of mixed microbial communities. In: Akkermans ADL, van Elsas JD, de Bruijn FJ, editors. Molecular microbial ecology methods. New York: Springer; 1996. pp. 1–23. [Google Scholar]
- Nikolova S, Tzevetkov Y, Najdenski H, Vesselinova AV. Isolation of pathogenic yersiniae from wild animals in Bulgaria. J Vet Med B. 2001;48:203–209. doi: 10.1046/j.1439-0450.2001.00448.x. [DOI] [PubMed] [Google Scholar]
- Oliveira M, Monteiro JL, Santos V, Carneiro C, Nunes SF, Vilela CL. Identification and antimicrobial resistance profile of gram-positive cocci from skin microbiota of timorese river buffalo (Bubalus bubalis) Proc Leibniz Inst Zoo Wildl Res. 2007;7:25–36. [Google Scholar]
- Quinn PJ, Carter ME, Markey B, Carter GR. Clinical veterinary microbiology. Edinburg: Mosby; 1994. p. 648. [Google Scholar]
- Santos-Reis M, Trindade A, Beja PR. Situation et état des recherches sur la loutre au Portugal. Cah “Ethnologia”. 1995;15:1–14. [Google Scholar]
- Santos-Reis M, Ferreira JP, Pedroso N, Baltazar C, Matos H, Pereira I, Grilo C, Sales-Luís T, Santos MJ, Cândido AT, Sousa I, Rodrigues M (2003) Projectos de Monitorização de Mamíferos. Monitorização de Carnívoros. Relatório Final. 2 Fase de Monitorização. Programa de Minimização para o Património Natural da área de regolfo de Alqueva e Pedrógão. Centro de Biologia Ambiental (FCUL)/Centro de Estudos da Avifauna Ibérica (CEAI), p 207
- Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Simpson VR. Veterinary advances in the investigation of wildlife diseases in Britain. Res Vet. Sci. 2000;69:11–16. doi: 10.1053/rvsc.2000.0384. [DOI] [PubMed] [Google Scholar]
- Smith WA, Mazet JAK, Hirsh DC. Salmonella in California wildlife species: prevalence in rehabilitation centers and characterization of isolates. J Zoo Wildl Med. 2002;33:228–235. doi: 10.1638/1042-7260(2002)033[0228:SICWSP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Trindade A, Farinha N, Florêncio E. A distribuição da lontra Lutra lutra em Portugal: situação em 1995. Estudos de Biologia e Conservação da Natureza do Instituto para a Conservação da Natureza. 1998;28:1–121. [Google Scholar]


