Abstract
The 3C-like protease (3CLpro) of severe acute respiratory syndrome associated coronavirus (SARS-CoV) is vital for SARS-CoV replication and is a promising drug target. Recombinant 3CLpro was expressed in Pichia pastoris GS115 as a 42 kDa protein that displayed a K m of 15 ± 2 μM with Dabcyl-KTSAVLQSGFRKME-Edans as substrate. Purified 3CLpro was used for inhibition and kinetic assays with seven flavonoid compounds. The IC50 of six flavonoid compounds were 47–381 μM. Quercetin, epigallocatechin gallate and gallocatechin gallate (GCG) displayed good inhibition toward 3CLpro with IC50 values of 73, 73 and 47 μM, respectively. GCG showed a competitive inhibition pattern with K i value of 25 ± 1.7 μM. In molecular docking experiments, GCG displayed a binding energy of −14 kcal mol−1 to the active site of 3CLpro and the galloyl moiety at 3-OH position was required for 3CLpro inhibition activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s10529-011-0845-8) contains supplementary material, which is available to authorized users.
Keywords: Flavonoid, Molecular docking, Pichia pastoris, Protease, Severe acute respiratory syndrome (SARS)
Introduction
In 2002, the first reported outbreak of severe acute respiratory syndrome (SARS) occurred in Guangdong province, China. It rapidly spread to over 32 countries in Asia, North America, and Europe. The mortality rate was approximately 10%, according to World Health Organization data. The causative agent of SARS is a novel human coronavirus (CoV), designated SARS-CoV. An efficient therapy and a vaccine are not currently available (Xu et al. 2005). SARS-CoV is an enveloped positive single-stranded RNA virus (Rota et al. 2003) that encodes two proteases for proteolytic processing: a papain-like cysteine protease (PLP2pro) and a chymotrypsin-like cysteine protease (3C-like protease; 3CLpro) located in the non-structural protein regions nsp3 and nsp5, respectively. Since 3CLpro is essential for the viral life cycle, it is an attractive target for the development of antiviral drugs directed against SARS-CoV and other CoV infections (Grum-Tokars et al. 2008).
Flavonoids are a large group of naturally occurring phenolic compounds ubiquitously distributed in the plant kingdom. Over 4,000 varieties of flavonoids have been identified (de Groot and Rauen 1998). Flavonoids can act as potent antioxidants, and display anti-inflammatory, antiallergic, antihemorrhagic, antimutagenic, antineoplastic, and hepatoprotective activities (Tapas et al. 2008; de Groot and Rauen 1998). In addition, flavonoids inhibit the catalytic activities of a great variety of enzymes, including hexokinase, phospholipase C, protein kinase C, α-glucosidase, and α-amylase. (Tadera et al. 2006). Some flavonoid compounds such as quercetin, quercetin derivatives, catechin, epicatechin, epicatechin gallate and epigallocatechin gallate inhibit SARS-3CLpro expressed in Escherichia coli (Chen et al. 2005, 2006; Yi et al. 2004). However, there has been no report on the high inhibition activity of gallocatechin gallate (GCG) against SARS-3CLpro, or the structure–activity relationship activity among the aforementioned flavonoid compounds.
Herein, we report on the expression of SARS-3CLpro in Pichia pastoris GS115, its’ in vitro inhibition by seven flavonoid compounds belonging to four groups of flavonoids (flavonol, flavanonol, isoflavone, and flavan-3-ol), and the structure–activity relationship between them. The detailed mechanism of GCG inhibition was investigated by enzyme kinetic and molecular docking studies.
Materials and methods
Preparation of recombinant 3CLpro
The procedures for construction, transformation, and screening for the catalytic domain of SARS 3CLpro were according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). The gene encoding SARS 3CLpro polyprotein (amino acid residues 3,241–3,546, GenBank accession no. AY274119) (Benson et al. 2011) was optimized by replacing rare codons with high-frequency codons, which were selected on the basis of the findings for codons usage in Pichia pastoris (Supplementary Fig. 1), and were synthesized and cloned in the pUC57 vector (pUC57-3CL) by a custom gene synthesis service (GenScript, Piscataway, NJ, USA). The 3CLpro gene was isolated from the vector pUC57-3CL by cutting with EcoRI and NotI, and was subcloned into the EcoRI/NotI restriction sites of the expression vector pPICZαA. The novel construct was named pPICZαA-3CLpro and transformed into E. coli DH5α (Promega, Madison, WI, USA) using a standard heat shock method. Transformants harbouring pPICZαA-3CLpro were selected from LB agar low salt medium [1% (w/v) Tryptone, 0.5% (w/v) yeast extract, 0.5% NaCl, pH 7.5] containing 25 μg Zeocin ml−1 (Invitrogen). Plasmid pPICZαA-3CLpro was amplified in E. coli DH5α, linearized by SacI digestion, and transformed into P. pastoris GS115 by a modified LiCl method. pPICZαA vector was also linearized by SacI digestion and transformed into P. pastoris GS115 as a negative control strain. Screening for positive clones was done by PCR with two sets of primers (Bioneer, Deajeon, Korea): set one contained 3CLpro primers [3CL-F (5′-GTGGATTCAGAAAAATGGCC-3′) and 3CL-R (5′-CCGCCTGAAAAGTAACTCCT-3′)] and set two contained α-factor and 3AOX1 primers. The PCR conditions were 94°C for 5 min, followed by 25 cycles of 94°C for 1 min, 53°C for 30 s, 72°C for 1 min, and a final step of 72°C for 5 min. PCR was conducted using PCRmix and the PCR product was analyzed by agarose gel electrophoresis.
Recombinant 3CLpro was expressed according to the manufacturer’s instructions (Invitrogen). Large-scale expression of 3CLpro was carried out in a 10 l fermenter with 4 l BMMY medium. The fermentation conditions were 28°C, pH 6.0, 350 rpm, and aeration at 1 vvm. Methanol was added as described above. Protein expression by pPICZαA transformed into P. pastoris GS115 was included as a negative control. The yeast was separated from the broth by centrifugation at 8,000×g for 15 min. The pellet was discarded and the supernatant was exchanged by 20 mM Tris/HCl buffer (pH 7.5) using a Millipore membrane. The supernatant was used for ammonium sulphate fractionation (from 0 to 85%). The obtained proteins were dissolved in 20 mM Tris/HCl buffer (pH 7.5) and dialyzed against 20 mM Tris/HCl buffer (pH 7.5). The detection of recombinant 3CLpro in the culture supernatant was by 12% SDS-PAGE and western blot of the electrotransferred proteins according to to the manufacturer’s instructions (GE Healthcare, Buckinghamshire, UK). Activity of the recombinant enzyme was detected as described below.
The proteolytic activity of 3CLpro was measured using a fluorescence resonance energy transfer (FRET)-based assay with a substrate labeled with 5-[(2′-aminoethyl)-amino]naphthelenesulfonic acid (Edans) and 4-[{4-(dimehtylamino)phenyl}azo]benzoic acid (Dabcyl) as the energy transfer pair (Bachem, Bubendorf, Switzerland). The Dabcyl-KTSAVLQSGFRKME-Edans fluorogenic peptide was used as the substrate and the enhanced fluorescence due to cleavage of this substrate catalyzed by the protease was monitored at 538 nm with an excitation wavelength of 355 nm using a fluorescence plate reader. The reaction mixture contained 3 μg 3CLpro protease and 20 μM fluorogenic substrate in 20 mM Tris/HCl buffer (pH 7.5) (Grum-Tokars et al. 2008). The plates were analyzed at 25°C with continuous monitoring of fluorescence for 25 min, with recording of relative fluorescence units (RFUs) using a SpectraMax Gemini XPS apparatus (Molecular Devices, Sunnyvale, CA, USA) with excitation and fluorescence emission wavelengths of 355 and 538 nm, respectively. Kinetic parameters of recombinant 3CLpro were obtained using 12.5–100 μM FRET peptides in the fluorescent assay with an 18 min measurement period. Reaction responses were linear within this time. The (K m) value was calculated from a Lineweaver–Burk using the SigmaPlot program (Systat Software, San Diego, CA, USA).
Inhibition assay
Quercetin, daidzein, puerarin, epigallocatechin (EGC), epigallocatechin gallate (EGCG), and gallocatechin gallate (GCG) were purchased from Sigma–Aldrich and ampelopsin (AMPLS) was purchased from ZR chemicals (Shanghai, China). Primarily inhibitory activities of flavonoid compounds were determined by measuring the remaining activity of 3CLpro at 200 μM inhibitors. GCG, EGCG, and EGC were dissolved in water; quercetin, puerarin, daidzein, and AMPLS were dissolved in dimethylsulfoxide (DMSO) as 10 mM stock solutions. The enzyme reaction digest (100 μl) was composed of 3 μg enzyme, 20 μM FRET substrate, 200 μM of each flavonoid compound, and 20 mM Tris/HCl buffer (pH 7.5). Reactions were run for 18 min at 25°C with continuous monitoring of fluorescence. The inhibition was calculated using following formula (1): % inhibition = 100 − remaining activity (%) where the remaining activity (%) = [(S − S o)/(C − C o)] × 100 (1), where C is the fluorescence of the control (enzyme, buffer, and substrate) after 18 min incubation, C o is the fluorescence of the control at time zero, S is the fluorescence of the tested samples (enzyme, tested sample solution, buffer and substrate) after 18 min incubation, and S o is the fluorescence of the tested samples at time zero. The 50% inhibitory concentration (IC50) was defined as the concentration of 3CLpro inhibitor necessary to reduce 3CLpro activity by 50% relative to a reaction mixture containing 3CLpro enzyme but no inhibitor. Inhibitor kinetic studies were performed for GCG, which was the best inhibitor against 3CLpro. The method was similar to those used in a kinetic study of the recombinant enzyme, except for the use of multiple concentrations of the inhibitor (0–60 μM) and variable concentrations of substrate (5–15 μM). The type of inhibition was determined using Lineweaver–Burk plots and a Dixon plot (1/v as a function of inhibitor concentration, [I]) and kinetic parameters (K i) was calculated using the SigmaPlot program.
Docking of flavonoid compounds with 3CLpro
The three-dimensional structure of 3CLpro was retrieved from the Protein Data Bank [http://www.pdb.org, accession code 2ZU5]. N-[(benzyloxy)carbonyl]-O-tert-butyl-l-threonyl-N-[(1R)-4-cyclopropyl-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl} butyl]- l-leucinamide (ZU5) was located in the active site of 2ZU5 (Lee et al. 2009). All water molecules, co-crystal ligand ZU5 were removed and the structure information containing only the amino acid residues of the 3CLpro enzyme was used for docking. Docking files were prepared using AutoDockTools software (Sanner et al. 1996). For the protein molecules, polar hydrogen atoms were added and nonpolar hydrogen atoms were merged. Kollman charges and solvation parameters were assigned by default. The three-dimensional atomic coordinates of GCG were generated by the Corina program (Molecular Networks GmbH, Erlangen, Germany), Gasteiger charges were added and nonpolar hydrogen atoms were merged. The grid box, with grid spacing of 0.375 Å and dimensions of 60 × 60 × 60 points along the x, y, and z axes, was centered on the macromolecule. AutoDock version 3.0.5 software using the Lamarckian genetic algorithm (LGA) was used for the computational molecular docking simulation of flexible small molecules to rigid proteins with ligand and rigid proteins (Morris et al. 1998). Important docking parameters for the Lamarckian genetic algorithm were a population size of 250 individuals, maximum of 5 million energy evaluations, maximum of 27,000 generations, mutation rate of 0.02, crossover rate of 0.80, and 100 docking runs (each docking job produced 100 docked conformations). The probability of performing a local search on an individual in the population was set to 0.06 and the maximum number of iterations per local search was set to 300. The conformation with the lowest docked energy was chosen from the most populated cluster and was put through to the next stage. The hydrogen bond (H-bond) interaction between 3CLpro and GCG was identified by Ligplot software (Wallace et al. 1995).
Results and discussion
Recombinant 3CLpro enzyme preparation
The 918 bp gene encoding 3CLpro (amino acids 3,241–3,546) from human SARS-CoV was cloned into the pPICZαA expression vector (pPICZαA-3CLpro) and the pPICZαA-3CLpro plasmid was linearized by SacI digestion that was further incorporated into the AOX1 locus of Pichia pastoris. Seven colonies were identified as 3CLpro-positive by PCR (Supplementary Fig 2). A single clone was selected for expression; the recombinant 3CLpro was resolved electrophoretically as a band of approx. 42 kDa. To confirm that the clone secreted 3CLpro, western blotting was performed using the anti-His antibody; 3CLpro was apparent as the same band upon SDS-PAGE and the control did not show this band on Western blot (Supplementary Fig 3).
Large-scale expression of 3CLpro was carried out in a 10 l fermenter with 4 l BMMY medium. 3CLpro was secreted and the amount of 3CLpro protein was increased as the induction time increased (Fig. 1a) as confirmed by western blot analysis (Fig. 1b). After 4 days induction, the RFU (as representative of enzyme activity) was increased 2.4-times compared with 3 days induction. Electrophoretic analysis of the 3CLpro fraction obtained using ammonium sulfate gave 80% purity (Supplementary Fig 4). The yields of each step in the procedure are summarized in Supplementary Table 1. No appreciable glycosylation was observed for 3CLpro even after endoglycosidase H treatment.
Fig. 1.
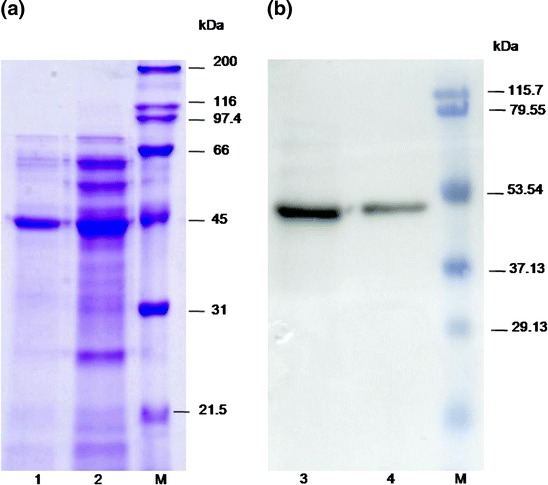
SDS-PAGE and Western blot results of recombinant 3CLpro after 3 and 4 days induction during fermentor culture. Lane M: marker; lanes 1 and 2: SDS-PAGE after 3 days (lane 1) and 4 days (lane 2); lanes 3 and 4: Western blot conducted after 3 days (lane 4) and 4 days (lane 3)
To calculate the kinetic parameters of the purified 3CLpro, enzyme activity was analyzed with fluorescent substrate from 12.5 to 100 μM. The Km value was 15 ± 1 μM (Supplementary Fig 5). The K m value of the purified 3CLpro was similar to the K m value of 3CLpro expressed in E. coli (K m of 17 ± 4 μM) (Kuo et al. 2004).
3CLpro inhibition by flavonoid compounds
Seven flavonoid compounds belonging to four groups of flavonoids (flavonol, flavanonol, isoflavone, and flavan-3-ol) (Fig. 2) were evaluated for their inhibitory activity against 3CLpro expressed from P. pastoris GS115. Table 1 shows the inhibitory activity of each flavonoid against the 3CLpro at 200 μM. Quercetin, EGCG and GCG inhibited more than 80% of the activity of recombinant 3CLpro; AMPLS, puerarin, and daidzein inhibited more than 30% activity of recombinant 3CLpro; and EGC inhibited only 5% of the activity of recombinant 3CLpro. Six compounds displayed an IC50 ranging from 47 to 381 μM (Table 1). GCG showed the best inhibition against recombinant 3CLpro with IC50 value of 47 ± 0.9 μM, and so was used for the further analysis of inhibition mode. Both Lineweaver–Burk and Dixon plots were used. As shown in Fig. 3a, GCG exhibited competitive inhibition toward 3CLpro because the Lineweaver–Burk plot of 1/v versus 1/[S] resulted in a family of straight lines with the same y-axis intercept. The K i value of GCG was determined to be 25 ± 1.7 μM from the common x-axis intercept of lines on the corresponding Dixon plot (Fig. 3b).
Fig. 2.
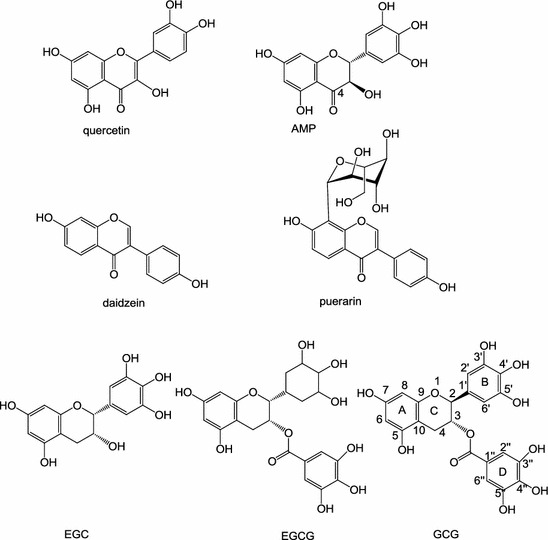
Molecular structures of the seven flavonoids
Table 1.
Inhibitory activity of flavonoid compounds against 3CLpro
| Compound | Inhibitiona (%) | IC50 (μM) | Docking score (kcal mol−1) |
|---|---|---|---|
| AMPLS | 34 | 364 ± 8.7 | −9.9 |
| Quercetin | 82 | 73 ± 4 | −10.2 |
| Puerarin | 33 | 381 ± 12.5 | −11.3 |
| Daidzein | 34 | 351 ± 2.9 | −8.6 |
| EGC | 5.4 | ND | −9.3 |
| EGCG | 85 | 73 ± 2 | −11.7 |
| GCG | 91 | 47 ± 0.9 | −14.1 |
ND not determined, GCG gallocatechin gallate, EGCG epigallocatechin gallate, EGC epigallocatechin, AMPLS ampelopsin
aInhibition by 200 μM
Fig. 3.

Lineweaver-Burk plot (a) and Dixon plot (b) analyses for the inhibition of 3CLpro by GCG. The kinetic constants, K m and K i, were calculated using linear regression analysis. a GCG concentration 0 μM (filled circle), 20 μM (open circle), 30 μM (filled inverted triangle), 40 μM (open triangle), 50 μM (filled square), 60 μM (open square). b FRET substrate concentrations 5 μM (filled circle), 7.5 μM (open circle), 10 μM (filled inverted triangle), and 15 μM (triangle)
Molecular docking on 3CLpro
In order to get a better comprehension of the molecular recognition process between 3CLpro and antioxidant compounds, docking experiments were performed using the crystal structure of 3CLpro (2ZU5). Autodock 3.0.5 was used to carry out docking simulations. The free binding energy of flavonoid compounds is shown in Table 1. Among them, GCG displayed the lowest free binding energy (−14 kcal mol−1). The binding between GCG and active site pocket of 3CLpro is shown in Fig. 4a. To elucidate the interaction of 3CLpro with GCG, the potential hydrophobic and H-bond interactions between amino acid residues in the active site pockets of 3CLpro and GCG were investigated using the Ligplot program. Figure 4b depicts the details of the specific interactions between GCG and 3CLpro. Carbon atoms of GCG interacted hydrophobically with His41, Cys145, Met165, Glu166, Asp187, Arg188, and Gln189 of 3CLpro. GCG formed seven hydrogen bonds with residues in the catalytic binding pocket of 3CLpro. The O atom of the main chain carboxyl group of Glu166 formed an H-bond with the O33 atom of the 5-hydroxyl group of the A ring with a distance of 2.56 Å. The O atom of hydroxyl group of aromatic side chain of Tyr54 formed a H-bond with the O30 atom of the 4′-hydroxyl group of the ring B at 2.7 Å. The O29 atom of the 5′-hydroxyl group of ring B accepted a H-bond from the N atom of the imidazol group of His41with a distance 2.72 Å. The O28 atom of the 3″-hydroxyl group of the galloyl group accepted a H-bond with the N atom from the imidazol group of His163 with a distance of 2.96 Å. The O26 atom of the 4″-hydroxyl group of the galloyl group has two H-bonds: one with the carboxyl group of Leu141 and another one with the carboxyl group of Ser144 at 2.90 and 2.49 Å, respectively. The N atom of the amino group of Gly143 donated a H-bond with an O27 atom of the 5″-hydroxyl group of the galloyl group with a distance of 3.35 Å.
Fig. 4.
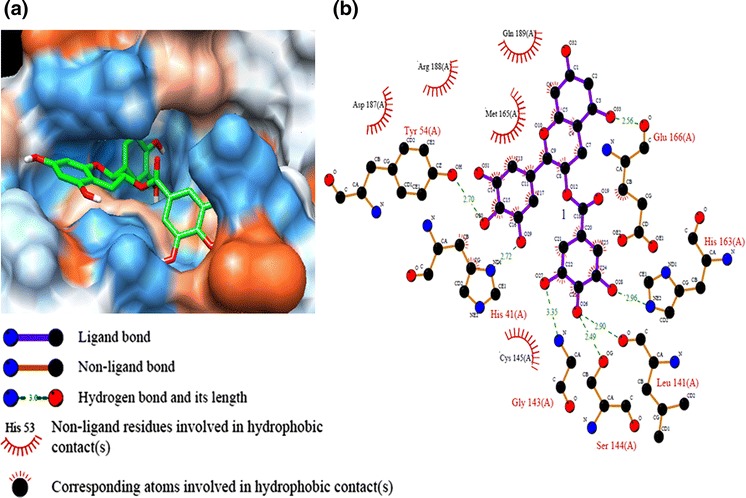
Computational docking and hydrophobic and hydrogen bond interactions of GCG with amino acid residues in the active site of 3CLpro. a Comparison of binding modes of GCG (green) in the active site pocket of 3CLpro. b Hydrophobic and H-bond interactions between GCG and amino acid residues in the active site of 3CLpro. H-bond interactions are represented by green dashed lines (Red, oxygen; cornflower blue, nitrogen; black, carbon)
Structural activity relationships of flavonoid compounds
In this study, we compared the inhibition activity of AMPLS, EGC, EGCG, and GCG containing the same B-ring at 200 μM (Table 1). The decreasing order of the inhibitory activity was EGC < AMPLS < EGCG < GCG. EGCG and GCG have a galloyl moiety at the 3-OH position, which is absent in the other catechins used in this study. EGCG and GCG displayed stronger 3CLpro inhibitory activity than those of EGC and AMPLS. GCG (2S, 3R type), which is a C-2 epimeric isomer of EGCG (2R, 3R type), showed 1.5-times higher 3CLpro inhibitory activity than that of EGCG. Molecular docking simulation was used to calculate the binding of GCG to the 3CLpro active site. GCG bound at the substrate-binding pocket of 3CLpro with numerous hydrophobic and hydrogen bond interactions. The galloyl group from GCG was important for GCG binding to 3CLpro active site pocket because it has four hydrogen bond interactions with Leu141, Gly143, Ser144, and His163 (Fig. 4b). The effect of B-ring and hydroxyl group substitution on the B-ring for the inhibitory activity was evaluated. Daidzein and puerarin, which lack the B-ring, showed little inhibition of 3CLpro. The 3CLpro inhibitory activity of AMLSP was 4.96-times lower than that of quercetin. This confirmed that the addition of an OH group at 5′-position of the B ring decreased the 3CLpro inhibitory activity. The effect of the structures of the A and C rings on the inhibitory activity was evaluated. Since AMPLS, which lacks the 2,3-double bonds in the C-ring, showed lower inhibitory activity than that of quercetin, 2,3-double bonds are probably crucially influential to the inhibitory activity. EGC lacking the C(4)=O in the C-ring, C(2)=C(3), containing 5′-OH group and lacking galloyl moiety showed the lowest inhibitory activity compared to that of AMPLS, daidzein, puerarin, quercetin, EGCG, or GCG.
Conclusions
An important goal of the present study was to understand the inhibition by seven flavonoid compounds belonging to four groups of flavonoids (flavonol, flavanonol, isoflavone, and flavan-3-ol) against 3CLpro of SARS-CoV. The active extracellular 3CLpro was successfully expressed and purified in P. pastoris GS115. Among the investigated flavonoids, GCG was the best inhibitor against 3CLpro by an in vitro assay. The structure and inhibition activity relationship among seven flavonoid compounds was also investigated. GCG showed numerous hydrophobic and H-bonds interaction with amino acid residues in the active site pocket of 3CLpro.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was partially supported by 21C Frontier Microbial Genomics and the Applications Center Program.
References
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CN, Lin CPC, Huang KK, Chen WC, Hsieh HP, Liang PH, Hsu JTA. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3, 3′-digallate (TF3) Evid Based Compliment Altern Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li J, Luo C, Liu H, Xu W, Chen G, Liew OW, Zhu W, Puah CM, Shen X, Jiang H. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): structure–activity relationship studies reveal salient pharmacophore features. Bioorg Med Chem. 2006;14:8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot H, Rauen U. Tissue injury by reactive oxygen species and the protective effects of flavonoids. Fund Clin Pharmacol. 1998;12(3):249–255. doi: 10.1111/j.1472-8206.1998.tb00951.x. [DOI] [PubMed] [Google Scholar]
- Grum-Tokars V, Ratia K, Begaye A, Baker SC, Mesecar AD. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CJ, Chi YH, Hsu JT, Liang PH. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem Biophys Res Commun. 2004;318:862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, Shih SR, Liang PH, Wang AHJ. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Tadera K, Minami Y, Takamatsu K, Matsuoka T. Inhibition of alpha-glucosidase and alpha-amylase by flavonoids. J Nutr Sci Vitam. 2006;52:149–153. doi: 10.3177/jnsv.52.149. [DOI] [PubMed] [Google Scholar]
- Tapas AR, Sakarkar DM, Kakde RB. Flavonoids as nutraceuticals: a review. Trop J Pharm Res. 2008;7:1089–1099. doi: 10.4314/tjpr.v7i3.14693. [DOI] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. Ligplot—a program to generate schematic diagrams of protein ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Xu T, Ooi A, Lee HC, Wilmouth R, Liu DX, Lescar J. Structure of the SARS coronavirus main proteinase as an active C-2 crystallographic dimer. Acta Crystallogr F. 2005;61:964–966. doi: 10.1107/S1744309105033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Li Z, Yuan K, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


