Abstract
Objectives
Viral oncoproteins are ideal targets in therapeutic vaccines for functional inhibition of human papillomaviruses (HPVs). Herein, we designed the peptide constructs derived from E5 and E7 oncoproteins of high-risk HPV types 16, 18, 31 and 45 using the bioinformatics tools and investigated their potency in mice.
Results
The framework of the combined in silico/in vivo analysis included (1) to determine physicochemical properties of the designed constructs, (2) to identify potential IFN-γ-inducing epitopes, (3) to assess allergenicity, (4) to recognize linear and discontinuous B cell epitopes using modeling and validation of 3D structure of the designed constructs, and (5) to evaluate immune responses and tumor growth in vivo. Our in silico data determined high potency of the HPV16,18,31,45 E5 and HPV16,18,31,45 E7 peptides for trigger B- and T-cell responses, and IFN-γ secretion. In vivo study indicated that the mixture of E5 and E7 immunodominant peptides from four types of high-risk HPV could induce Th1 immune response, and protect completely mice against TC-1 tumor cells.
Conclusion
Generally, the combined in silico/in vivo approaches showed the ability of the designed E5 and E7 peptide constructs from four major high-risk HPV types for development of therapeutic vaccines.
Electronic supplementary material
The online version of this article (10.1007/s10529-020-02792-6) contains supplementary material, which is available to authorized users.
Keywords: Human papillomavirus, Oncoprotein, Peptide vaccine, Bioinformatics analysis, In vivo studies
Introduction
With more than 200 genotypes, human papillomavirus (HPV) is the main reason for cervical cancer, and most common sexually transmitted viruses in the world (Bruggmann et al. 2018). Based on the annual report of the International Agency for Research on Cancer evaluation (IARC), twelve types of HPVs (i.e., 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59) were classified as high-risk types. Among high-risk HPV types, HPV 16, 18, 31 and 45 had the highest prevalence among women with cervical cancer in the world (Bruggmann et al. 2018). HPV genome has an 8 kb circular double strand DNA that encodes eight open reading frames (ORFs) consisting of the early and late genes. Early proteins (E1, E2, E4, E5, E6 and E7) play the vital role in cell cycle, signaling pathway, genome replication and transcription. In contrast, late proteins (L1 and L2) are involved in capsid formation (Liu et al. 2017; Graham 2010). Nowadays, three commercially HPV prophylactic vaccines are available, but none of them show a therapeutic effect on existing HPV infection and related cancers (Bharadwaj et al. 2009; Joura et al. 2015). Thus, due to high prevalence of HPV infections, development of an effective therapeutic vaccine is indispensable. Moreover, HPV E5, E6 and E7 proteins are ideal targets for generating a therapeutic vaccine due to their important roles in tumor pathogenesis, cellular transformation (degradation of p53 and pRB) and virus replication (Hoppe-Seyler et al. 2018; Ganguly 2012). Up to now, several therapeutic vaccines have been developed based on viral oncoproteins as ideal targets including live vector-based vaccines, bacterial or viral vectors-based vaccines, DNA and RNA vectors-based vaccines, peptide and protein-based vaccines, and whole cell-based vaccines. However, each strategy showed some advantages and disadvantages (Yang et al. 2016) [8]. Thus, it is important to find effective and safe methods in therapeutic vaccine design (e.g., increasing their immunogenicity) (Gomez-Gutierrez et al. 2007; Cassetti et al. 2004; Reinis et al. 2010). Among different approaches, peptide vaccines were known as a key strategy because of various advantages such as high selectivity and sensitivity, easy production and cost-effective. A valuable peptide vaccine should be composed of immunodominant T- and B-cell epitopes that reduce immune responses against self-antigens (i.e., autoimmunity). Therefore, a main step is the interaction between epitopes with major histocompatibility complex (MHC) (Li et al. 2014). Bioinformatics tools have been developed for different aspects of immunological features such as epitope prediction and mapping, molecular modeling and structural vaccinology (Sirskyj et al. 2011). These tools can predict the highly immunogenic epitopes in a short time with high specificity, and can be used for development of an effective vaccine (Jurtz et al. 2017). In this study, to design a novel therapeutic vaccine against major high risk HPV types 16, 18, 31 and 45, we used both sequence-based and structural vaccinology immunoinformatics tools. The E7 and E5 proteins of each HPV were considered to design peptide constructs. After preparation of constructs using bioinformatics analysis, their potency was assessed by in vivo studies (i.e., induction of immune responses and tumor eradication in mice).
Materials and methods
Selection of immunodominant epitopes
Based on our previous study (Panahi et al. 2018), eight epitopes for both E5 (SAFRCFIVYIIFVYIPLFLIHTHARF-HPV16, SPATAFTVYVFCFLL-HPV18, YVVFIYIPLFVIHTHASF-HPV31, QSVYVCAFAWLLVF-HPV45) and E7 (GQAEPDRAHYNIVTF-HPV16, SSADDLRAFQQLFL-HPV18, GQAEPDTSNYNIVTF-HPV31 and TLQEIVLHLEPQNELDPVDLL-HPV45) proteins were selected. According to bioinformatics analysis, these epitopes had the highest scores in important parameters such as binding affinity between peptide and MHC, MHC-I processing, peptide-MHC docking, and population coverage.
Physicochemical properties of the designed constructs
The physicochemical properties of the designed constructs such as molecular weight, theoretical pI, negatively and positively charged residues, estimated half-time, instability index and solvent accessibility were determined by ProtParam (https://web.expasy.org/protparam/) tools, Predictprotein (https://www.predictprotein.org) and Scratch servers (https://scratch.proteomics.ics.uci.edu/).
Prediction of interferon-gamma inducing epitopes
To determine the ability of the selected epitopes to induce interferon-gamma, IFNepitope server (https://crdd.osdd.net/raghava/ifnepitope/predict.php) was used. In this study, we used Motif and SVM hybrid algorithms (accuracy of 81.39%) and IFN-gamma versus Non IFN-gamma model for prediction (Dhanda et al. 2013).
Allergenicity assessment
Proteins have significant roles in inducing an allergenic reaction, thus their potential allergenicity should be determined especially in vaccine development.
The allergenicity of the selected epitopes was calculated by PA3P server (https://lpa.saogabriel.unipampa.edu.br:8080/pa3p/pa3p/pa3p.jsp) using AFDS-motif, Allergen online (6aa and 80 wordmatch) algorithms. The specificity of these methods was 88.1% (AFDS), 92.88% (80aa) and 95.43% (6aa) (Chrysostomou and Seker 2014).
D structure modeling
The 3D structures of the designed constructs were generated by I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). Based on CASP13 experiments, I-TASSER server was ranked as No 1 server for predicting 3D structure of proteins. I-TASSER server used hierarchical approach for prediction of protein structure including (a) Identification of structural template from PDB by LOMETS, (b) Fragments assembly, (c) Model selection, and (d) Functional annotation (Yang et al. 2015).
Refinement of 3D structures
Top 3D structure model obtained from I-TASSER was refined by GalaxyRefine 2 Server (https://galaxy.seoklab.org/cgi-bin/submit.cgi/type=REFINE2). GalaxyRefine2 performs iterative optimization with several geometric operators to increase accuracy of the initial model. This method utilizes global operators (e.g., anisotropic normal mode perturbation and secondary structure perturbation) and local operators (e.g., loop modeling and hybridization) as well as local error estimation and homolog structure information (Lee et al. 2018).
Validation of refined 3D structure
To validate and select the best model of a refined structure, all models of E5 and E7 obtained from GalaxyRefine2 server were analyzed by SAVE v5.0 server (https://servicesn.mbi.ucla.edu/SAVES/). SAVE v5.0 server used five different algorithms (ERRAT, Prove, Procheck, Ramachandran plot and whatcheck) to validate and check the stereochemical quality and atomic interaction of predicted 3D structure.
Prediction of linear and discontinuous antibody epitopes
To predict linear and discontinuous antibody epitopes, refined model was analyzed by ElliPro server (https://tools.iedb.org/ellipro/help/). ElliPro server used modified Thornton’s method along with residue clustering algorithms. Epitope prediction parameters (minimum score and maximum distance) were set to default values (0.5 and 6) (Ponomarenko et al. 2008).
Peptide synthesis
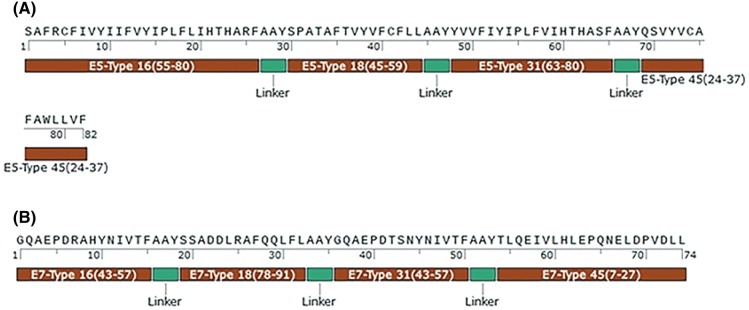
For evaluation of immunity, two peptide constructs (Fig. 1) were synthesized by BioMatik Corporation (Canada) with more than 75% purity. The synthesized peptides were prepared in salt form (acetate salt) under cold chain situation. According to the company instructions, the peptides were received as lyophilized form, dissolved in water (1 mg/mL), and stored in − 20 °C.
Fig. 1.
a E5 peptide construct, b E7 peptide construct. The AAY sequence (alanine/alanine/tyrosine) was used as a proteolytic linker
Mice immunization
Inbred C57BL/6 female mice, 5–7 week old, were obtained from the breeding stocks maintained at the Pasteur Institute of Iran. Mice were maintained under specific pathogen-free conditions and all procedures were performed according to approved protocols by Pasteur Institute of Iran (national guideline) for scientific purposes. Five groups of six mice were selected. Mice were immunized on days 0, 14, and 28 with 20 µg of the peptide constructs (HPV16,18,31,45 E7 peptide and HPV16,18,31,45 E5 peptide) in Phosphate-buffered saline (PBS1X, Sigma). The peptides were emulsified with Montanide ISA720 (GSK Company) at the ratio of 70:30 (v/v, oil: aqueous phase). Montanide ISA 720 was made of natural metabolizable non-mineral oil and a highly refined emulsifier from the mannide mono-oleate family which is rapidly metabolized and eliminated (Aucouturier et al. 2002).
Prophylactic effects
Inbred C57BL/6 female mice were subcutaneously injected in the right footpad with different regimens three times with a 2-week interval as indicated in Table 1. 2 weeks after the last immunization, mice were subcutaneously challenged in the right flank with 1 × 105 TC-1 tumor cells, and then monitored for tumor growth and the percentage of tumor-free mice (i.e., survival rates) by palpation twice a week. At each time point, tumor size was determined by measuring the smallest diameter (a) and the biggest diameter (b) by caliper. Tumor volume was calculated using the formula: V = (a2b)/2 (Li et al. 2006). The TC-1 cancerous cell line (ATCC number: CRL-2785) was derived from primary lung epithelial cells of C57BL/6 mice co-transformed with HPV16 E6, HPV16 E7 and ras oncogenes (Ji et al. 1998, 1999). These cells were cultured in RPMI 1640 (Sigma, Germany) supplemented with 5% heat-inactivated fetal bovin serum (FBS, Sigma, Germany), 2 mM l-glutamine (Sigma, Germany) and 40 µg/mL gentamicin (Sigma, Germany), and incubated at 37 °C in 5% CO2. On the day of tumor challenge, TC-1 cells were harvested by trypsinization, washed with PBS1X, counted and finally resuspended in 500 µL of PBS1X. The schematic model of immunization program was shown in Supplementary 1.
Table 1.
Immunization program in prophylactic study
| Groups | Vaccine modality | Priming | Booster 1 (2 weeks after priming) | Booster 2 (2 weeks after booster 1) | Challenge with TC-1 (2 weeks after booster 2) |
|---|---|---|---|---|---|
| G1 | Peptide/peptide/peptide | E5 + montanide | E5 + Montanide | E5 + montanide | TC-1 |
| G2 | Peptide/peptide/peptide | E7 + montanide | E7 + Montanide | E7 + montanide | TC-1 |
| G3 | Peptide/peptide/peptide | E5 + E7 + montanide | E5 + E7 + Montanide | E5 + E7 + montanide | TC-1 |
| G4 | Control | PBS | PBS | PBS | TC-1 |
| G5 | Control | Montanide | Montanide | Montanide | TC-1 |
Antibody responses
The mice in prophylactic study were bled from retro-orbital (after anesthesia using intraperitoneal injection of Ketamine (87.5 mg/kg)/Xylazine (12.5 mg/kg) cocktail: 0.1 mL/20 g mouse) at 2 weeks after the second booster and then, the sera were pooled for each group. We used an indirect ELISA to detect the production of antibodies to E5, E7 and E5 + E7 peptides. Briefly, a 96-well flat-bottom ELISA plate (Greiner, Germany) was coated overnight at 4◦C with 100 µL of each antigen [i.e., E7 peptide (10 µg/mL), E5 peptide (10 µg/mL) or E7 + E5 peptides (10 µg/mL)] diluted in PBS1X (pH = 7.2, Sigma, Germany). Then, the plate was rinsed with washing buffer (0.5% (v/v) Tween-20 in PBS1X), incubated with blocking buffer (1% BSA in PBS1X, Sigma, Germany) for 2 h at 37◦C. The pooled sera were diluted 1:100 in dilution buffer (0.5% (v/v) Tween-20 in blocking buffer), added to the plate, and incubated for 2 h at 37 °C. After rinsing with washing buffer, the plate was incubated with horseradish peroxidase-conjugated goat anti-mouse IgG1, IgG2a, IgG2b or total IgG antibodies (diluted 1:10,000 in 1% BSA/PBS-Tween, Sigma, Germany) for 2 h at 37 °C. Detection was done with 100 µL of 3,3′,5,5′-Tetramethylbenzidine or TMB (Sigma, Germany) as the substrate followed by incubation for 10 min at room temperature. The enzyme reaction was stopped by 0.5 M H2SO4 (Merck, Germany) and the absorbance was measured at 450 nm.
Cytokine assay
Three mice of each group in prophylactic study were sacrificed randomly after anesthesia before TC-1 challenge. The spleens were removed, homogenized and the red blood cell-depleted pooled splenocytes (2 × 106 cells/mL) were cultured in 48-well plates (Nunc, Germany) containing RPMI medium supplemented with 10% heat-inactivated FBS for 72 h in the presence of 10 μg/mL of the E7 peptide, the E5 peptide or the E7 + E5 peptides, and 5 μg/mL of concanavalin A (ConA) as positive control. The supernatants (100 µL/well) were harvested and the generation of IFN-γ, IL-5 and IL-10 cytokines was measured with the sandwich-based ELISA method using a Maptek ELISA kit according to the manufacturer’s instructions. All data were represented as mean ± SD for each sample.
Granzyme B assay
The P815 target cells (T; 2 × 104 cells/well) were seeded in triplicate into U-bottomed, 96-well plates (Nunc, Germany) and incubated with the E7 + E5 peptides (~ 30 μg/mL) for 24 h. The part of the prepared splenocytes (Effector cells: E) in cytokine assay was added to the target cells at E:T ratio of 100:1. The target and effector cells were co-cultured in complete RPMI medium supplemented with 10% heat-inactivated FBS at 37 °C and 5% CO2 under humidified conditions. After 6 h incubation, the microplates were centrifuged at 250×g for 5 min at 4 °C and the supernatants were harvested to assess the concentration of Granzyme B by ELISA (eBioscience) according to the manufacturer’s instruction.
Therapeutic effects
At first, 1 × 105 TC-1 tumor cells were subcutaneously inoculated in the right flank of 3 mice in each group. 1 week after TC-1 inoculation, C57BL/6 mice were subcutaneously injected in the right footpad with 20 µg of the E7 + E5 peptide regimen (G1), and PBS (G2, control). Two booster doses were injected 2 weeks after the first injection with a 2-week interval. Tumor growth was monitored twice a week by inspection and palpation for two months.
Statistical analysis
The differences between the control and test groups were assessed using one-way ANONA (Graph-pad Prism 5.0, GraphPad Software). Survival rate or the percentage of tumor-free mice was evaluated using the log-rank (Mantel–Cox) test. The value of p < 0.05 was considered statistically significant. Two independent experiments were performed to obtain reproducibility. Two replicates were used to evaluate immune responses in each experiment.
Results
Physicochemical properties of the designed constructs
Based on our previous study (Panahi et al. 2018), two different peptide constructs were designed (Fig. 1). According to bioinformatics prediction, these epitopes had the highest degree of immunogenicity, binding affinity with MHC molecules, and population coverage. Herein, ProtParam, Predictprotein and Scratch servers evaluated physicochemical properties of these constructs. The results obtained from these servers were summarized in Table 2. Our data showed that the E5 and E7 peptide constructs were stable (with the instability index (II) of 21.71 and 29.00, respectively) and soluble (with the probability of 0.711 and 0.88, respectively). High-performance liquid chromatography (HPLC) and mass spectrometry (MS) reports of the synthesized peptides were shown in Supplementary 2 and 3. The purity of the E5 and E7 peptides was 77.84% and 75.51%, respectively.
Table 2.
Physicochemical properties of the designed E5 and E7 epitopes
| Construct | Molecular weight (kDa) | Theoretical PI | Disulphide bond | Positive charge residue | Negative charge residue | Solubility |
|---|---|---|---|---|---|---|
| E5 | 9.5 | 8.52 | Between 41 and 74 (34 amino acid length) | 2 | 0 | Soluble |
| E7 | 8.2 | 4.02 | None | 2 | 11 | Soluble |
Prediction of allergenictiy and IFN-γ-inducing epitopes
Since the generation of IFN-γ plays a key role in the control of HPV infection, the E5 and E7 candidate peptides were analyzed by IFNepitope server. The results showed that SPATAFTVYVFCFLL (E5-type 18), QSVYVCAFAWLLVF (E5-type 45) and TLQEIVLHLEPQNELDPVDLL (E7-type 45) peptides had the best IFN-γ-inducing scores (Table 3). Allergenicity analysis showed that the candidate peptides were not allergen.
Table 3.
IFN-γ inducing scores of the designed E5 and E7 epitopes
| Protein | Epitope sequence | IFN-γ inducing score* | Allergenicity |
|---|---|---|---|
| E5 | SPATAFTVYVFCFLLA | + 0.57 (positive) | None |
| QSVYVCAFAWLLVF | + 0.25 (positive) | None | |
| SAFRCFIVYIIFVYIPLFLIHTHARF | − 2.80 | None | |
| VVFIYIPLFVIHTHASF | − 4.00 | None | |
| E7 | TLQEIVLHLEPQNELDPVDLL | + 1.55 (positive) | None |
| SSADDLRAFQQLFL | − 1.00 | None | |
| GQAEPDTSNYNIVTF | − 1.31 | None | |
| GQAEPDRAHYNIVTF | − 0.99 | None |
*Higher rates show more potent epitopes for inducing IFN-γ
Modeling of 3D structures
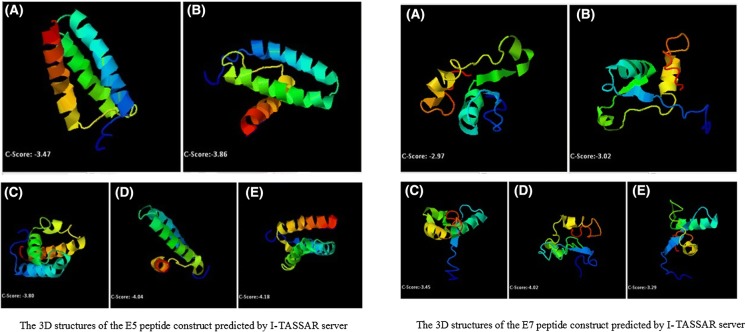
For prediction of the 3D structure of the designed epitopes, I-TASSER server was used. At first, I-TASSER generated a structural conformation called decoys. Then, to select the final models, I-TASSER used the SPICKER program to cluster all the decoys based on the pair-wise structure similarity. Finally, I-TASSER reported the top five models which correspond to the five largest structure clusters. The assurance of each model was calculated by C-score that was based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations. C-score is usually in the range of [− 5, 2], where a C-score of a higher value indicated a model with a higher confidence. C-scores of the best model for the E5 and E7 peptide constructs were − 3.47 and − 2.97, respectively. Figure 2 showed the best 3D structures of the E5 and E7 peptide constructs and their C-score.
Fig. 2.
Left picture The 3D structures of the E5 peptide construct predicted by I-TASSAR server. The higher value of C-score indicates a model with a higher confidence. In this case, model A with C-score of -3.47 has the highest score among the predicted structures; Right picture The 3D structures of the E7 peptide construct predicted by I-TASSAR server. The higher value of C-score indicates a model with a higher confidence. In this case, model A with C-score of -2.97 has the highest score among the predicted structures
Refinement and validations
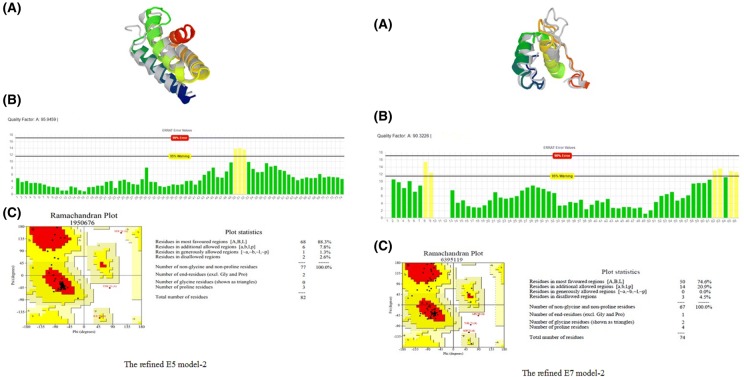
For each construct, the best 3D structures obtained from the I-TASSER server were submitted separately to GalaxyRefine2 server. After refinement analysis, the top refined model was entered to the next step which was validation of the 3D structures. According to the results of SAVE5.0 server, E5-model No. 1 and E7-model No. 2 refined structures had the highest quality factors (95.94 and 90.32, respectively) which were selected for further analysis. Figure 3 showed the best-refined model and its characteristics.
Fig. 3.
Left picturea The refined E5 model-2 generated by GalaxyRefine 2 Server. b ERRAT error values chart and quality factor of E7 Model-2. c Ramachadran plot of E7-model-2 shows 88.3% of residues in the most favoured regions; Right picturea The refined E7 model-2 generated by GalaxyRefine 2 Server, b ERRAT error values chart and quality factor of E7 Model-2. c Ramachadran plot of E7-model-2 shows 74.6% of residues in the most favoured regions
Prediction of linear and discontinuous antibody epitopes
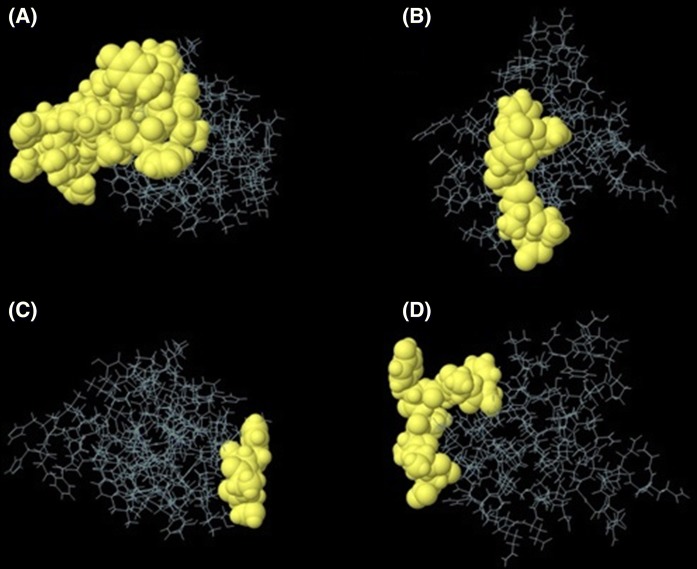
Linear antibody epitopes (or B cell epitopes) can be predicted using sequence-based algorithms and mainly based on amino acid properties such as charge and hydrophobicity. Nevertheless, prediction of discontinuous epitopes needs information of 3D structure of a protein. In this study, the selected refined model was analyzed by Ellipro server to predict potential linear and discontinuous B cell epitopes. In E5 peptide construct, one linear epitope (VFIYIPLFVIHTHASF) and four discontinuous epitopes were identified. In E7 peptide construct, four linear epitopes (FAAYSSAD, GQAEPDRAHY, EPDTSNYNIVT and LHLEPQNELDP), and five discontinuous epitopes had the highest epitope prediction scores (Table 4). Figure 4 indicated an example of the 3D structure of putative B cell epitopes in the E5 and E7 peptide constructs.
Table 4.
Linear and discontinuous B cell epitopes of the designed vaccine constructs
| Construct | Epitope | Score* |
|---|---|---|
| Linear B cell epitopes | ||
| E5 | VFIYIPLFVIHTHASF | 0.718 |
| E7 | FAAYSSAD | 0.720 |
| GQAEPDRAHY | 0.716 | |
| EPDTSNYNIVT | 0.638 | |
| LHLEPQNELDP | 0.577 | |
| Construct | Epitope | Score |
|---|---|---|
| Discontinuous B Cell epitopes | ||
| E5 | A:S1, A:A2, A:F3 | 0.789 |
| A:A27, A:A28, A:V73, A:F76, A:A77, A:L79, A:L80, A:V81, A:F82 | 0.774 | |
| A:V13, A:Y47, A:V50, A:F51, A:I52, A:I54, A:P55, A:L56, A:F57, A:V58, A:I59, A:H60, A:T61, A:H62, A:A63 | 0.706 | |
| A:S30, A:P31, A:A32, A:T33 | 0.646 | |
| E7 | A:F15, A:A16, A:A17, A:Y18, A:S19, A:S20, A:A21, A:D22 | 0.720 |
| A:G1, A:Q2, A:A3, A:E4, A:P5, A:D6, A:R7, A:A8, A:H9, A:Y10 | 0.716 | |
| A:Y53, A:E57, A:L60, A:H61 | 0.703 | |
| A:E63, A:P64, A:Q65, A:E67, A:L68, A:D69, A:P70 | 0.608 | |
| A:Q37, A:A38, A:E39, A:P40, A:D41, A:T42, A:S43, A:N44, A:Y45, A:N46, A:I47, A:V48, A:T49 | 0.583 | |
*Higher rates show better quality of epitope identification
Fig. 4.
a 3D view of “VFIYIPLFVIHTHASF” linear B cell epitope on the E5 peptide construct. b 3D view of “FAAYSSAD” linear B cell epitope on the E7 peptide construct. c 3D view of “A:S1, A:A2, A:F3” discontinuous B cell epitope on the E5 peptide construct. d 3D view of “A:F15, A:A16, A:A17, A:Y18, A:S19, A:S20, A:A21, A:D22” discontinuous B cell epitope on the E7 peptide construct. Yellow bubbles show the position of B cell epitopes on the E5 and E7 peptide constructs
Evaluation of antibody responses
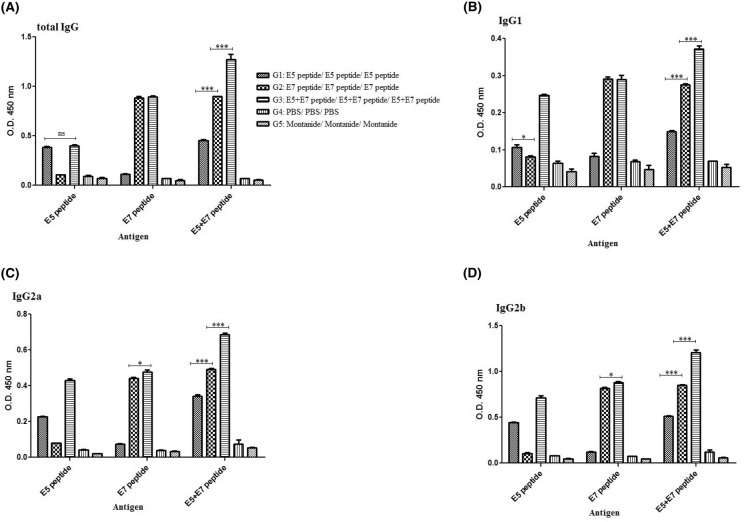
The levels of total IgG and the related subclasses (IgG1, IgG2a and IgG2b) were assessed against the E7, E5 and E7 + E5 peptides in various groups (Fig. 5a–d). Our data indicated that the highest levels of total IgG, IgG1, IgG2a and IgG2b was observed in the sera of mice vaccinated with the mixture of E7 + E5 peptides (G3) among all groups (p < 0.05, Fig. 5). Moreover, the E7 peptide could further increase the secretion of total IgG, IgG1, IgG2a and IgG2b as compared to the E5 peptide (p < 0.05). It was interesting that the peptide constructs could significantly induce IgG2b as compared to IgG2a and IgG1 production (p < 0.05, Fig. 5). Indeed, our data indicated the mixture of IgG1, IgG2a and IgG2b with high intensity toward IgG2a and IgG2b responses in all groups especially the group receiving the mixture of E7 + E5 peptides (G3). No significant anti-E5 and anti-E7 antibody responses could be detected in the sera of control groups. Indeed, all test groups showed significant antibody responses against antigens as compared to control groups (p < 0.05). Thus, the seroreactivities were completely E5 or E7 antigen-specific responses in C57BL/6 mice.
Fig. 5.
Antibody responses against the peptides as antigens in different regimens: total IgG (a), IgG1 (b), IgG2a (c), IgG2b (d); all analyses were performed in duplicate for each sample. The results were shown as mean absorbance at 450 nm ± SD. *p < 0.05; **p < 0.01, ***p < 0.001, ns non-significant (p > 0.05)
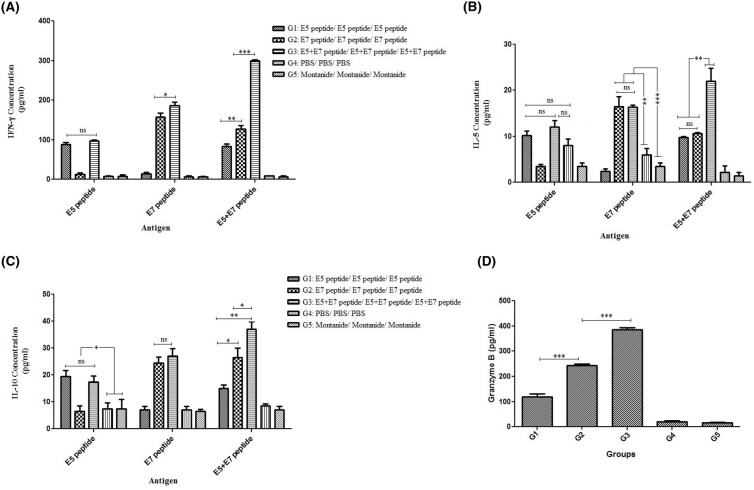
Secretion of cytokines
The cytokine results for the pooled splenocytes of three mice in each group indicated that the levels of the E7-specific and E7 + E5-specific IFN-γ secretion in the group vaccinated with E5 + E7 peptide prime/E5 + E7 peptide boost (G3) regimen were significantly higher than those in other groups (p < 0.05, Fig. 6a). The E5-specific IFN-γ secretion did not show any considerable difference between groups immunized with the E5 peptide alone (G1) and the E5 + E7 peptides (G3, p > 0.05). All mice immunization with different modalities effectively enhanced the levels of IFN-γ against both peptides as compared to control groups (p < 0.05). The data indicated that the E7 peptide was more effective than the E5 peptide for IL-5 secretion. Among all the test groups, the group immunized with the E7 + E5 peptide (G3) showed a significant IL-5 response (~ 22 pg/mL) against the E7 + E5 peptides compared to other groups (~ 10 pg/mL, p < 0.05, Fig. 6b). Moreover, all the test groups demonstrated the E5, E7, E5 + E7-specific IL-10 secretion higher than control groups (p < 0.05, Fig. 6c). Our data also showed that the ratios of IFN-γ/IL-5 and IFN-γ/IL-10 were higher in all test groups especially group receiving the E5 + E7 peptides (the mean of IFN-gamma/IL-5 ratios were about 8, 10 and 14, and the mean of IFN-gamma/IL-10 ratios were about 5, 7 and 8 against the E5, E7 and E5 + E7 peptides, respectively); thus they could significantly activate the Th1 cellular immune response.
Fig. 6.
Secretion of cytokines in immunized groups with various formulations: The levels of IFN-γ (a), IL-5 (b), IL-10 (c) secretion were determined in the supernatants using ELISA as mean absorbance at 450 nm ± SD for each sample. All analyses were performed in duplicate for each sample; Granzyme B secretion measured by ELISA (d): All analyses were performed in triplicate for each sample. The results represent mean values calculated from triplicate samples as well as the standard deviation (SD) as error bars. *p < 0.05; **p < 0.01, ***p < 0.001, ns non-significant (p > 0.05)
Granzyme B secretion
Two weeks after the last immunization, Granzyme B secretion in each sample was measured by ELISA. The data showed that group immunized with the E5 + E7 peptide produced significantly higher concentrations of Granzyme B than other groups (p < 0.05, Fig. 6d). Moreover, group immunized with the E7 peptide could significantly enhance Granzyme B secretion compared to group immunized with the E5 peptide (p < 0.05) indicating more potency of E7 peptide than E5 peptide in secretion of Granzyme B as a possible indicator of cytotoxic T lymphocyte (CTL) activity.
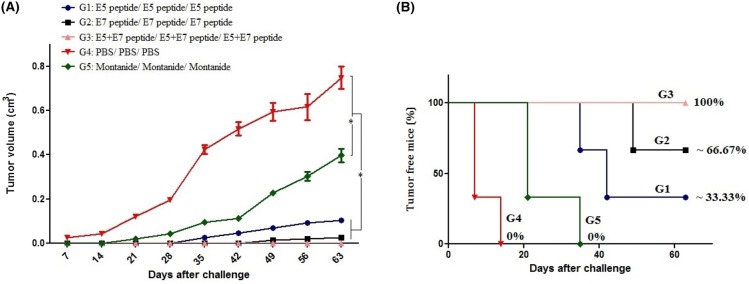
Mice protection against E7-expressing tumor cells
For evaluation of vaccine potency, tumor growth was assessed in all groups. As shown in Fig. 7a, tumor growth was significantly reduced in all test groups especially group receiving the E5 + E7 peptides (G3) as compared to control groups (G4 and G5). Indeed, all mice in control groups developed tumor growth on approximately 7–35 days. As indicated in Fig. 7b, immunization with the E5 + E7 peptides (G3) could protect completely all mice from tumor growth (100% tumor-free mice) compared to the E5 peptide (G1: ~ 33.33%), the E7 peptide (G2: ~ 66.67%) and also control groups (G4 or G5: 0%).
Fig. 7.
Prophylactic studies against TC-1 tumor cells: Five groups of C57BL/6 mice were vaccinated with different regimens three times with a 2-week interval and then challenged with 1 × 105 TC-1 in the right flank 2 weeks after second booster. Tumor volumes were measured twice a week (a). The percentage of tumor-free mice was determined over time in various groups (b)
Mice treatment against tumor growth
Mice with pre-established E7-expressing tumors were injected with the mixture of E5 + E7 peptides (G1), and PBS (G2), 1 week after tumor inoculation. Tumor size and animal survival were monitored for 60 days following the challenge. Our data showed that treatment with the E5 + E7 peptide prime/E5 + E7 peptide boost regimen effectively suppressed the tumor growth in mice more than 60 days (100% tumor-free mice, Supplementary 4).
Discussion
Therapeutic vaccine is an attractive strategy to stimulate the immune system against cancer-associated HPV high-risk types. A therapeutic vaccine should target HPV antigens that can be constitutively expressed in HPV-associated malignancies (Schiller et al. 2008). Because of HPV E5, E6 and E7 roles in tumor pathogenesis, cellular transformation and virus replication, they are ideal targets for generating a therapeutic vaccine (Kumar et al. 2015; Chabeda et al. 2018). Peptide vaccines are safe, stable and easy to produce but they are restricted to a specific major histocompatibility complex (MHC). Another issue is the low immunogenicity of peptide-based vaccines. Therefore, bioinformatics tools were used to solve these problems and design a multi-epitope vaccine. Due to the advantageous use of bioinformatics tools, many studies used these tools to design a peptide-based vaccine against a hypervariable virus such as human immunodeficiency virus (HIV) (Li et al. 2013; Khairkhah et al. 2018), dengue virus (Chakraborty et al. 2010) and coronavirus (Oany et al. 2014). Numeral previous studies applied bioinformatics tools for epitope prediction and therapeutic vaccine design based on HPV E5 and E7 proteins. In 2015, Kumar et al. found 11 potent epitopes in HPV16-E5 protein for MHC-I based on immunogenicity scores (Kumar et al. 2015). In 2016, Singh et al. studied E1, E2, E6 and E7 protein sequences collected from all high-risk HPV types to identify cross-clade immunodominant regions. They introduced 14 peptides (9 to 43 amino acids) which can be used as potent regions in therapeutic vaccines (Singh et al. 2016). Tsang et al. analyzed HPV16 E6 and E7 proteins and found six immunogenic epitopes, as well (Tsang et al. 2017).
While the oncogenic activities of E6 and E7 were well known, the role of E5 is still rather unclear. The recent studies have indicated the important role of E5 oncoprotein in cell transformation, tumourigenesis, and immune modulation (Venuti et al. 2011). The researchers showed that HPV16 E5 oncoprotein was expressed in early stage of carcinogenesis and could be a target of immunotherapy (Paolini et al. 2017). On the other hand, the presence of E5 viral transcripts could be a major marker of active viral infection, and subsequently a target of immunotherapy. HPV16 E5 was highly expressed in HPV16-positive oropharyngeal cancer (OPC) patients (Taberna et al. 2018). Generally, most studies have focused on E7 oncoprotein, because it is more abundantly expressed and better characterized immunologically. Furthermore, its sequence is more conserved than that of the E6 gene (Mahdavi and Monk 2005). In this study, we considered E5 protein as a novel target and E7 protein as a major target in vaccine design. Based on our previous study (Panahi et al. 2018) and to overcome the MHC limitation, the long overlapping peptide constructs containing HPV16,18,31,45 E5 and E7 epitopes were designed. These epitopes had the highest scores in important parameters such as binding affinity between peptide and MHC, MHC-I processing, peptide-MHC docking and population coverage. In addition to previous analyses, we used structural bioinformatics tools to determine (1) physicochemical properties of the designed constructs, (2) potential IFN-γ-inducing epitopes, (4) allergenicity, and (5) linear and discontinuous B cell epitopes by modeling and validation of 3D structure of the designed constructs. Subsequently, for in vivo analysis, the candidate E5 and E7 peptide constructs were synthesized, and their immunological responses and anti-tumor effects were evaluated in mice. The physicochemical properties of the E5 and E7 peptide constructs showed that they are stable, soluble and non-allergen. In addition, the E5 peptide construct had 2 positive charge residues and the E7 peptide construct consisted of 2 positive and 11 negative charge residues. On the other hand, IFN-γ has a key role in intracellular immunity against HPV infection (Day et al. 2017). Our data showed that SPATAFTVYVFCFLLA (E5-type 18) and TLQEIVLHLEPQNELDPVDLL (E7-type 45) had the highest scores for stimulation of IFN-gamma against HPVs. In the case of 3D modeling of the E5 and E7 peptide constructs, I-TASSER server was selected to predict the protein 3D structure. The accuracy of the selected models was evaluated by C-score (significance of threading template alignments and the convergence parameters of the structure assembly simulations). C-scores of the best models for E5 and E7 peptides were − 3.47 and − 2.97, respectively. These data showed that the accuracy of E7 is greater than E5. Based on the Ramachandran plots and ERRAT scores of the refined models, the quality of the predicted 3D constructs was improved after refinement (Fig. 3) leading to a higher quality of final models. Final E5 and E7 3D models were selected as input for B-cell linear and discontinuous epitope predictions. In the current study, Ellipro server analyzed 3D models and found one linear epitope (VFIYIPLFVIHTHASF) and four discontinuous epitopes in the E5 peptide construct, and four linear epitopes (FAAYSSAD, GQAEPDRAHY, EPDTSNYNIVT and LHLEPQNELDP) and five discontinuous epitopes in the E7 peptide construct. These data indicated the ability of the designed constructs for induction of B-cell responses. In addition, our previous bioinformatics analysis (containing molecular docking and sequence-based approaches) showed that these epitopes had high potency for inducing CTL and T helper cell responses. These epitopes represented high population coverage (~ 99%) among different area of the world. After bioinformatics analysis, immunological assays of the designed constructs were studied in mice. Indeed, we compared the potency of each peptide as individual (HPV16,18,31,45 E5 polytope peptide or HPV16,18,31,45 E7 polytope peptide) with the combined regimen (HPV16,18,31,45 E5 polytope peptide plus HPV16,18,31,45 E7 polytope peptide) as a candidate antigen designed by bioinformatics analyses. For enhancement of peptide immunity, Montanide ISA720 was used as a commercial adjuvant. Our results showed that the E5/E7 peptide-specific immunity in mice who received the E5 + E7 peptides (G3) resulted in the highest levels of IFN-γ and IgG2b secretion among all groups. A study indicated that the recombinant lipidated HPVE7 induced a high level of IgG2b/IgG1, a Th1-biased immune response, and protective immunity in mice (Huang et al. 2012). Regarding to our observations in protective and therapeutic studies, this regimen (the E5 + E7 peptides) could completely confer protection against TC-1 tumor cell-challenged mice depending on stimulation of CD4+ T cell-dominated Th1 responses as well as Granzyme B secretion. Some experimental studies demonstrated the immunogenicity of different identified antigenic peptides of HPV16/18 E6 and E7 proteins. For instance, a study showed that the intranasal administration of the combined HPV16 peptide vaccine [E744–62 peptide (QAEPDRAHYNIVTFCCKCD); E749–57 peptide (RAHVYNIVTIF); E643–57 peptide (QLLRREVYDFAFRDL); and E649–58 peptide (VYDFAFRDLC)) with α4-1BB and αCTLA-4 antibodies produced efficient therapeutic effects and high safety against orally implanted mEER tumors (Dorta-Estremera et al. 2018). Moreover, the use of CpG motif (ODN1826: 5′-TCCATGACGTTCCTGACGTT-3′) as an adjuvant with E7 peptide-based immunotherapy (The H-2b-restricted E7 CTL epitope: RAHYNIVTF) led to reduce the tumor growth (Gendron et al. 2006). Other study showed that the conjugation of HPV16 E7 long peptide (E743-77: GQAEPDRAHYNIVTFCCKCDSTLRLCVQSTHVDIR) to ultra-small polymeric nanoparticles could enhance the antitumor effects in different mouse models of HPV+ cancers (Galliverti et al. 2018). Van der Burg et al. reported the immunogenic epitopes of HPV16 E741–62 including a CTL epitope (QAEPDRAHY) (van der Burg et al. 2001; Jabbar et al. 2018). Our predicted HPV16 E7 epitope involved this reported epitope (GQAEPDRAHYNIVTF), as well. Currently, many reports indicated that long epitopes (containing 15-mers or longer) could induce an effective immune response as compared to short epitopes (Jabbar et al. 2018). According to these experimental reports, we designed two vaccine constructs with longer peptide epitopes (from 14 to 26 amino acid length). Up to now, most of the HPV therapeutic vaccines have been focused on HPV E6 and E7 oncogenes. However, these vaccines could not completely eradicate the lesions. Recently, HPV E5 oncogene attracted a special attention. Liao et al. showed that the injection of an E5 peptide along with CpG motif induced effective cellular immune responses and prolonged the survival time after tumor cell inoculation. This study demonstrated the importance of HPV16 E5 as a possible target for development of the therapeutic strategies against cervical cancer (Liao et al. 2013). Our study showed that the mixture of E5 and E7 immunodominant epitopes from four types of high risk HPV could generate high levels of IFN-γ, IgG2b and IgG2a against IL-5, IL-10 and IgG1 indicating the induction of Th1 immune responses as well as high Granzyme B secretion indicating CTL activity as compared to groups receiving E7 or E5 peptides, individually. Indeed, the mixture of E7 and E5 peptides could elicit higher immune responses and stronger anti-tumor effects (100% for the E7 + E5 peptides against 66.67% for the E7 peptide and 33.33% for the E5 peptide).
In general, the results of bioinformatics tools showed a higher level of IFN-γ secretion and B cell epitope prediction scores for E7 peptide versus E5 peptide (Tables 3 and 4). In this line, the in vivo experiment confirmed bioinformatics data by showing a higher level of IFN-γ secretion and total IgG for E7 peptide (Fig. 6). In addition, bioinformatics tools predict high solubility and molecular weight of peptides that was confirmed in in vitro analysis (Supplementary 2 and 3). However, it should be noted that in vivo experimental data indicated higher capacity of peptides for trigger immune system in the combined form (i.e., E5 + E7). Indeed, the bioinformatics prediction is a pointer of potential immunogenicity of peptide but it is not a criterion for function assignment. Therefore, bioinformatics analysis should be confirmed with experimental approaches.
Conclusion
In conclusion, we used both structural and sequence-based bioinformatics tools to predict efficient epitopes for development of a therapeutic vaccine against four prevalent high-risk HPV types in the world. The in vivo results supported the immunogenic potential of these predicted peptides especially in the combined form (i.e., the mixture of E5 and E7 peptides). Herein, E5 peptide could increase immune responses that non-specifically enhanced the E7 potency against tumor growth. However, there are limitations in the recent study such as the lack of tumor cell line expressing E5 oncoprotein. Development of a tumor cell line expressing E7 and E5 can be considered as an important approach in the next studies. Moreover, it is suitable for using the designed peptides in different modalities such as heterologous prime/boost strategy as well as the use of delivery systems for improvement of therapeutic vaccine in near future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors acknowledge the financial support by NIMAD “National Institute for Medical Research Development” for experimental works (Grant No. 963295).
Supporting information
Supplementary 1—The schematic model of immunization program in prophylactic study.
Supplementary 2—A) MS report of the E5 peptide construct, B) HPLC report of the E5 peptide construct.
Supplementary 3—A) MS report of the E7 peptide construct, B) HPLC report of the E7 peptide construct.
Supplementary 4—The percentage of tumor-free mice determined over time in groups receiving the E5+E7 peptide (G1) and PBS (G2) for therapeutic studies .
Author contributions
AB and AN conceived and designed the experiments. AN, HAP and AB reviewed all data, analyzed the results, and wrote the manuscript. AN and HAP performed the experiments. AB reviewed the manuscript and provided scientific advices.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All mice were maintained under pathogen-free conditions. The whole process was operated based on approval protocols and care of laboratory animals at Pasteur Institute of Iran and National Institute for Medical Research Development (Grant No. 963295).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aucouturier J, Dupuis L, Deville S, Ascarateil S, Ganne V. Montanide ISA 720 and 51: a new generation of water in oil emulsions as adjuvants for human vaccines. Expert Rev Vaccines. 2002;1(1):111–118. doi: 10.1586/14760584.1.1.111. [DOI] [PubMed] [Google Scholar]
- Bharadwaj M, Hussain S, Nasare V, Das BC. HPV & HPV vaccination: issues in developing countries. Indian J Med Res. 2009;130:327–333. [PubMed] [Google Scholar]
- Bruggmann D, Kayser L, Jaque J, Bundschuh M, Klingelhofer D, Groneberg DA. Human papilloma virus: Global research architecture assessed by density-equalizing mapping. Oncotarget. 2018;9:21965–21977. doi: 10.18632/oncotarget.25136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetti MC, McElhiney SP, Shahabi V, et al. Antitumor efficacy of Venezuelan equine encephalitis virus replicon particles encoding mutated HPV16 E6 and E7 genes. Vaccine. 2004;22:520–527. doi: 10.1016/j.vaccine.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Chabeda A, Yanez RJR, Lamprecht R, Meyers AE, Rybicki EP, Hitzeroth II. Therapeutic vaccines for high-risk HPV-associated diseases. Papillomavirus Res. 2018;5:46–58. doi: 10.1016/j.pvr.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Chakravorty R, Ahmed M, et al. A computational approach for identification of epitopes in dengue virus envelope protein: a step towards designing a universal dengue vaccine targeting endemic regions. In Silico Biol. 2010;10(6):235–246. doi: 10.3233/ISB-2010-0435. [DOI] [PubMed] [Google Scholar]
- Chrysostomou C, Seker H. Prediction of protein allergenicity based on signal-processing bioinformatics approach. Conf Proc IEEE Eng Med Biol Soc. 2014;2014:808–811. doi: 10.1109/EMBC.2014.6943714. [DOI] [PubMed] [Google Scholar]
- Day PM, Thompson CD, Lowy DR, Schiller JT. Interferon gamma prevents infectious entry of human papillomavirus 16 via an L2-dependent mechanism. J Virol. 2017;91:e00168-17. doi: 10.1128/JVI.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorta-Estremera S, Chin RL, Sierra G, et al. Mucosal HPV E6/E7 peptide vaccination in combination with immune checkpoint modulation induces regression of HPV+ oral cancers. Cancer Res. 2018;78:5327–5339. doi: 10.1158/0008-5472.CAN-18-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliverti G, Tichet M, Domingos-Pereira S, et al. Nanoparticle conjugation of human papillomavirus 16 E7-long peptides enhances therapeutic vaccine efficacy against solid tumors in mice. Cancer Immunol Res. 2018;6:1301–1313. doi: 10.1158/2326-6066.CIR-18-0166. [DOI] [PubMed] [Google Scholar]
- Ganguly N. Human papillomavirus-16 E5 protein: oncogenic role and therapeutic value. Cell Oncol. 2012;35:67–76. doi: 10.1007/s13402-011-0069-x. [DOI] [PubMed] [Google Scholar]
- Gendron KB, Rodriguez A, Sewell DA. Vaccination with human papillomavirus type16 E7 peptide with CpG oligonucleotides for prevention of tumor growth in mice. Arch Otolaryngol Head Neck Surg. 2006;132:327–332. doi: 10.1001/archotol.132.3.327. [DOI] [PubMed] [Google Scholar]
- Gomez-Gutierrez JG, Elpek KG, Montes de Oca-Luna R, et al. Vaccination with an adenoviral vector expressing calreticulin-human papillomavirus 16 E7 fusion protein eradicates E7 expressing established tumors in mice. Cancer Immunol Immunother. 2007;56:997–1007. doi: 10.1007/s00262-006-0247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SV. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5:1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158–168. doi: 10.1016/j.tim.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Huang CY, Chen JJW, Shen KY, et al. Recombinant lipidated HPV E7 induces a Th1-biased immune response and protective immunity against cervical cancer in a mouse model. PLoS ONE. 2012;7(7):e40970. doi: 10.1371/journal.pone.0040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar B, Rafique S, Salo-Ahen OMH, et al. Antigenic peptide prediction from E6 and E7 oncoproteins of HPV types 16 and 18 for therapeutic vaccine design using immunoinformatics and MD simulation analysis. Front Immunol. 2018;9:3000. doi: 10.3389/fimmu.2018.03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Chang EY, Lin KY, Kurman RJ, Pardoll DM, Wu TC. Antigen-specific immunotherapy for murine lung metastatic tumors expressing human papillomavirus type 16 E7 oncoprotein. Int J Cancer. 1998;78:41–45. doi: 10.1002/(sici)1097-0215. [DOI] [PubMed] [Google Scholar]
- Ji H, Wang TL, Chen CH, et al. Targeting human papillomavirus type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus type 16 E7-expressing tumors. Hum Gene Ther. 1999;10(17):2727–2740. doi: 10.1089/10430349950016474. [DOI] [PubMed] [Google Scholar]
- Joura EA, Giuliano AR, Iversen OE, et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N Engl J Med. 2015;372:711–723. doi: 10.1056/NEJMoa1405044. [DOI] [PubMed] [Google Scholar]
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairkhah N, Namvar A, Kardani K, Bolhassani A. Prediction of cross-clade HIV-1 T-cell epitopes using immunoinformatics analysis. Proteins. 2018;86:1284–1293. doi: 10.1002/prot.25609. [DOI] [PubMed] [Google Scholar]
- Kumar A, Yadav IS, Hussain S, Das BC, Bharadwaj M. Identification of immunotherapeutic epitope of E5 protein of human papillomavirus-16: An in silico approach. Biologicals. 2015;43:344–348. doi: 10.1016/j.biologicals.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Lee GR, Heo L, Seok C. Simultaneous refinement of inaccurate local regions and overall structure in the CASP12 protein model refinement experiment. Proteins. 2018;86:168–176. doi: 10.1002/prot.25404. [DOI] [PubMed] [Google Scholar]
- Li Y, Subjeck J, Yang G, Repasky E, Wang XY. Generation of anti-tumor immunity using mammalian heat shock protein 70 DNA vaccines for cancer immunotherapy. Vaccine. 2006;24:5360–5370. doi: 10.1016/j.vaccine.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Liang J, et al. Immune responses induced in HHD mice by multiepitope HIV vaccine based on cryptic epitope modification. Mol Biol Rep. 2013;40:2781–2787. doi: 10.1007/s11033-012-2202-y. [DOI] [PubMed] [Google Scholar]
- Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide vaccine: progress and challenges. Vaccines. 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SJ, Deng DR, Zeng D, et al. HPV16 E5 peptide vaccine in treatment of cervical cancer in vitro and in vivo. J Huazhong Univ Sci Technolog Med Sci. 2013;33:735–742. doi: 10.1007/s11596-013-1189-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pan Y, Gao W, Ke Y, Lu Z. Whole-genome analysis of human papillomavirus types 16, 18, and 58 isolated from cervical precancer and cancer samples in chinese women. Sci Rep. 2017;7:263. doi: 10.1038/s41598-017-00364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi A, Monk BJ. Vaccines against human papillomavirus and cervical cancer: promises and challenges. Oncologist. 2005;10:528–538. doi: 10.1634/theoncologist.10-7-528. [DOI] [PubMed] [Google Scholar]
- Oany AR, Emran AA, Jyoti TP. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Des Dev Ther. 2014;8:1139–1149. doi: 10.2147/DDDT.S67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahi HA, Bolhassani A, Javadi G, Noormohammadi Z. A comprehensive in silico analysis for identification of therapeutic epitopes in HPV16, 18, 31 and 45 oncoproteins. PLoS ONE. 2018;13:e0205933. doi: 10.1371/journal.pone.0205933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini F, Curzio G, Cordeiro MN, et al. HPV16 E5 oncoprotein is expressed in early stage carcinogenesis and can be a target of immunotherapy. Hum Vaccines Immunother. 2017;13(2):291–297. doi: 10.1080/21645515.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J, Bui HH, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinis M, Stepanek I, Simova J, et al. Induction of protective immunity against MHC class I-deficient, HPV16-associated tumours with peptide and dendritic cell-based vaccines. Int J Oncol. 2010;36:545–551. doi: 10.3892/ijo_00000528. [DOI] [PubMed] [Google Scholar]
- Schiller JT, Castellsague X, Villa LL, Hildesheim A. An update of prophylactic human papillomavirus L1 virus-like particle vaccine clinical trial results. Vaccine. 2008;26:K53–K61. doi: 10.1016/j.vaccine.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KP, Verma N, Akhoon BA, Bhatt V, Gupta SK, Smita S. Sequence-based approach for rapid identification of cross-clade CD8+ T-cell vaccine candidates from all high-risk HPV strains. J Biotech. 2016;6:39. doi: 10.1007/s13205-015-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirskyj D, Diaz-Mitoma F, Golshani A, Kumar A, Azizi A. Innovative bioinformatic approaches for developing peptide-based vaccines against hypervariable viruses. Immunol Cell Biol. 2011;89:81–89. doi: 10.1038/icb.2010.65. [DOI] [PubMed] [Google Scholar]
- Taberna M, Torres M, Alejo M, et al. The use of HPV16-E5, EGFR and pEGFR as prognostic biomarkers for oropharyngeal cancer patients. Front Oncol. 2018;8:589. doi: 10.3389/fonc.2018.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KY, Fantini M, Fernando RI, et al. Identification and characterization of enhancer agonist human cytotoxic T-cell epitopes of the human papillomavirus type 16 (HPV16) E6/E7. Vaccine. 2017;35:2605–2611. doi: 10.1016/j.vaccine.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg SH, Ressing ME, Kwappenberg KM, et al. Natural T-helper immunity against human papillomavirus type 16 (HPV16) E7-derived peptide epitopes in patients with HPV16-positive cervical lesions: identification of 3 human leukocyte antigen class II-restricted epitopes. Int J Cancer. 2001;91:612–618. doi: 10.1002/1097-0215. [DOI] [PubMed] [Google Scholar]
- Venuti A, Paolini F, Nasir L, et al. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer. 2011;10:140. doi: 10.1186/1476-4598-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Farmer E, Wu TC, Hung CF. Perspectives for therapeutic HPV vaccine development. J Biomed Sci. 2016;23(1):75. doi: 10.1186/s12929-016-0293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.