Abstract
The aim of this epidemiological study was to determine the prevalence of respiratory viruses, including new viruses, in hospitalised children in Austria. Two hundred fourteen nasopharyngeal samples from hospitalised children were tested for the presence of viruses using cell culture and PCR and/or viral antigen assays. The results revealed a parainfluenza virus 1 (PIV1) outbreak that ended right before the onset of the influenza season, with nearly no overlapping, moderate respiratory syncytial virus (RSV) activity, and only a few adenoviruses. Human metapneumovirus (hMPV) was present in 14.5% of the total samples but was detected in combination with other viruses in only five cases: with PIV1 in three cases and with RSV in two cases. There were no cases of dual infection with hMPV and flu or adenovirus. This suggests that hMPV alone is a leading cause of hospitalisation in children under 1 year of age. Interestingly, hMPV, in contrast to RSV, coincided with PIV1 but was absent during the community outbreak of the flu. Samples were also tested for Mimiviridae, a group of newly described DNA viruses that are similar to Legionella spp., replicate in water amoebae, and also have been found in alveolar cells. However, mimivirus was detected neither in respiratory samples nor in amoebae-containing water samples, indicating that this particular type of virus is either not abundant or does not contribute to paediatric respiratory illnesses.
Keywords: Influenza, Respiratory Syncytial Virus, Respiratory Syncytial Virus Infection, Dual Infection, Human Metapneumovirus
Introduction
The contribution of respiratory viruses to hospitalisation of children is often unclear. Viral emergence varies on a seasonal and annual basis, and the exact demographic characteristics of children affected are often unknown. Moreover, many viral infections are missed during routine laboratory diagnosis due to the poor quality of many clinical specimens, the lack of sensitivity of rapid antigen detection tests, and the prohibitively high cost of reverse transcriptase-PCR methods, which limits their use. The very high sensitivity of PCR methods is problematic because it is difficult to distinguish between residual genetic material from previous infections and the replicating viruses in an acute infection. Thus, nucleic acid detection methods, when used alone, may be difficult to interpret in cases of dual or multiple infections. Recent publications have shown that respiratory viruses may persist, without causing symptoms, for prolonged periods of time in the nasopharyngeal tract; in addition, noninfective viral nucleic acids from rhinovirus or respiratory syncytial virus (RSV) can be detected in humans or animal models for up to several months after infection [1, 2].
The recently described human metapneumovirus (hMPV) has been suggested to be less pathogenic than RSV, on the basis of hospitalisation rates and duration of symptoms of the patients affected [3, 4]. The impact of hMPV on the overall paediatric respiratory disease burden is still under investigation, and the role of hMPV in increasing the severity of RSV infection remains controversial [5, 6].
Another new virus family, the Mimiviridae, was first described in water amoebae obtained from a British cooling tower. These huge DNA viruses (400 nm diameter) were first though to be small bacteria and were thus named after their ability to “mimic” a microbe. Although there is no definitive evidence that they are pathogenic in humans, recent data suggests that they can be transmitted to humans because mimivirus DNA was found in broncheoalveolar specimens of ICU patients [7]. Moreover, serologic evidence of infection was observed in 5 of 26 (19.2%) ICU patients in the same study. We included this new virus in our preliminary study because there are currently no data available in paediatric patients.
Materials and methods
Origin of respiratory samples
A total of 214 nasopharyngeal aspirate samples were collected between the last week of September and 31 March at the local paediatrics department. Two hundred nine originated from nonimmunocompromised children who had been hospitalised for respiratory tract infections of different severity, and five originated from immunocompromised children. Of the immunocompromised children, two had undergone renal transplantation and one had received a heart transplant. Most of the children were admitted to hospital because of bronchiolitis, bronchitis, or pneumonia. Five children had malformities that involved the thorax or lungs or that necessitated oxygen therapy over longer periods of time. One child had cystic fibrosis.
Detection of viral antigen from respiratory specimens
For detection of RSV, the Pathfinder RSV antigen test (Bio-Rad, Hercules, CA, USA) was used. Influenza A and B antigen was detected with Flu-Directigen FluA+B (Becton Dickinson, Franklin Lakes, NJ, USA). To detect adenovirus, PCR was used in addition to the Premier Adenoclone kit (Meridian, Bioscience, Cincinnati, OH, USA).
Isolation of viruses
The human epithelial cell line HEp-2 (ATCC CCL-23) and the monkey kidney cell line MA-104 (ATCC CRL-2378) were used for culture of viruses. HEp-2 was preferred, since a recent study showed that these cells were superior to the rhesus monkey kidney cells (LLC-MK2) commonly used in previous studies for isolation of hMPV, especially if used in conjunction with a molecular method to detect hMPV, performed as a follow-up to cell culture [8]. HEp-2 laryngeal carcinoma cells, which, according to ATCC genetic analyses are contaminated by HeLa cells, also support the growth of adenoviruses, RSV, and some rhinoviruses, although rhinoviruses were not targeted in this study. The MA-104 line, which, according to recent genetic analysis at the ATCC, is a mixture of rhesus monkey kidney and African green monkey kidney cells, was chosen due its proven capability to grow parainfluenza viruses, SARS coronavirus, avian pneumoviruses, and a broad range of enteric viruses.
One millilitre of nasopharyngeal aspirates was filtered through sterile 0.44-μm membrane filters (Millipore, Billerica, MA, USA), inoculated into cell cultures, and allowed to stand for 1 day. Thereafter, the medium was removed and the cells cultured in DMEM with 2% foetal calf serum for 2 weeks for the HEp-2 cells and for 4 weeks for the MA-104 cells. Afterwards, culture tubes were frozen irrespective of any cytopathic effects observed.
Preparation of samples and controls
Two ml of supernatant from freeze–thawed cell cultures, which corresponds to approximately 5 × 105 cells, was processed with the QIAmp UltraSens virus kit (Qiagen, Valencia, CA, USA) and viral RNA/DNA eluted in 50 μl of H2O. Ten μl of the extracted RNA was reverse-transcribed using the SuperScript III Platinum RT kit (Invitrogen, Carlsbad, CA, USA) and random hexameres according to the manufacturer’s protocol. As positive controls, an enteric adenovirus isolate from stool (type 41) and parainfluenza virus types 1 (ATCC VR-94) and 3 (ATCC VR-93) were used. cDNA from hMPV was kindly provided by Dr. Barbara Huck, Freiburg, Germany.
PCR testing
Parainfluenza-virus- and adenovirus-specific PCR tests were performed in accordance with published protocols [9]. A PCR test for hMPV, which detects all hMPV lineages known thus far, has been described recently [10]. A PCR protocol suitable for detection of Mimiviridae in respiratory samples was kindly provided by D. Raoult, Marseille, France. To avoid specimen contamination during detection of such a rare pathogen, a “suicide PCR” protocol was used, which was described previously by the same group for detection of Yersinia pestis, the agent of medieval black death [11]. In the suicide PCR, primers are used only once, and there are no positive controls introduced into the laboratory. A negative test result is followed by a new test using other primers; a positive result is followed by sequencing. Although this procedure restricts sensitivity of detection of mimivirus to a certain extent, it provides absolute specificity by avoiding cross-contamination [7].
DNA sequencing
For typing of certain isolates of parainfluenza virus, hMPV, and adenovirus, DNA sequencing using an Applied Biosystems ABI 3100 automated sequencer (Applied Biosystems, Foster City, CA, USA) was used to further characterise the strains. The sequences obtained were compared to published sequences from the Genbank database using the BLAST search algorithm.
Results
Influenza virus
Only influenza A virus was detected in children younger than 3 years (mean age of infection, 14.6 months). The main influenza A peak ranged from mid-November to the end of February (Fig. 1). Two dual infections, one with RSV and one with adenovirus, were observed.
Fig. 1.
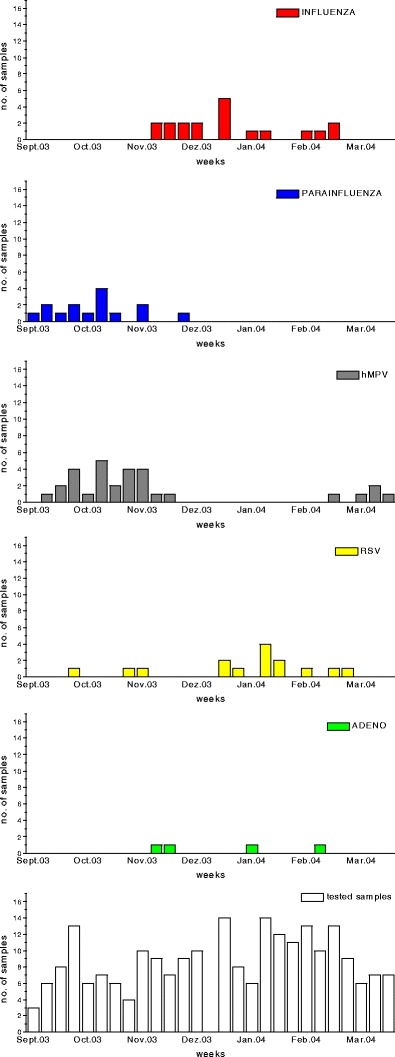
Graphical outline of respiratory samples that tested positive for viruses, with each week of detection depicted. Columns represent cumulative samples detected within 1 week, with the abscissae indicating the respective months from the last week of September to the end of March. The ordinates show the numbers of positive samples. Viruses investigated in this study are, from the top, influenza virus, parainfluenza virus, human metapneumovirus (hMPV), respiratory syncytial virus (RSV) and adenovirus (adeno). Mimivirus was also investigated, but none was detected. The total number of samples received/tested weekly is shown in the bottom graph (empty bars) for comparison
Parainfluenza virus
A seasonal peak from September to mid-November, which ended abruptly before the influenza A outbreak, was observed. Only a single case of PIV1 infection was detected within the main influenza A season (Fig. 1). Four samples from the PIV1 outbreak, including the sample from December, were confirmed by sequencing as having the PIV1 genotype. Most cases of parainfluenza virus infection occurred in children younger than 2 years (mean age of infection, 8.7 months) (Table 1). Two cases were observed in children older than 5 years: one in an ICU patient, and the other in a patient with lung malformation.
Table 1.
Mean age of children infected with influenza A virus, adenovirus, human metapneumovirus (hMPV), parainfluenza virus 1 (PIV1), and respiratory syncytial virus (RSV)
| Virus | Mean age of infected children (months) |
|---|---|
| Influenza A virus | 14.6 |
| Adenovirus | 12.2 |
| hMPV | 10.1 |
| PIV1 | 8.7 |
| RSV | 5.8 |
Adenovirus
Only four cases of adenovirus infection were detected in our hospitalised population: two were detected by PCR and two by the antigen test. All children with adenovirus infection were younger than 2 years (mean age of infection, 12.2 months) (Table 1). One dual infection of adenovirus type 2 (confirmed by sequencing) and influenza A occurred in a 10-month-old child.
Respiratory syncytial virus
No larger RSV epidemics were detected in hospitalised infants in our region during the time period studied (Fig. 1). Most cases of RSV infection occurred in children younger than 1 year (mean age of infection, 5.8 months), although three cases occurred in children older than 5 years (Table 1). Only three dual infections were observed, two with hMPV (1 of which occurred in a child undergoing respirator treatment) and one with influenza A virus.
Human metapneumovirus
Numerous isolates of hMPV were recovered from September to November, but the virus was almost absent during the influenza season. A small peak caused by isolates of the identical genotype was also observed in March (Fig. 1). In total, 31 of 214 (14.5%) samples were positive for hMPV, of which 29 originated from nonimmunocompromised children. Most cases occurred in children younger than 2 years (mean age of infection, 10.1 months), with more than 50% of the children affected being under 6 months of age (Table 2). Five dual infections were noted, three with PIV1 and two with RSV. No dual infections with influenza A virus or adenovirus were detected. One 15-year-old boy acquired a coinfection with RSV while recovering from enteroviral meningitis.
Table 2.
Age distribution of the 214 children in the study group and of the 29 hMPV-positive children
| Age range | No. of children | |
|---|---|---|
| All children in study | hMPV-positive children | |
| <6 months | 83 (39%) | 15 |
| 6 months to 1 year | 49 (23%) | 4 |
| 1–2 years | 43 (20%) | 4 |
| 2–4 years | 16 (7%) | 4 |
| >4 years | 23 (11%) | 2 |
Mimivirus
Mimivirus was not detected in any of the respiratory samples tested. Moreover, several amoeba-containing samples from water tanks in Austria as well as from Thailand all tested negative in the mimivirus-specific suicide PCR (data not shown).
Discussion
Since the clinical relevance of PCR findings in respiratory samples is not always clear due to detection of residual genetic material [1, 2], we employed a combination of cell culture and PCR to detect actively replicating respiratory viruses in this epidemiological investigation. The combination of HEp-2 and MA-104 cells for viral culture enabled detection of not only numerous hMPV and parainfluenza virus isolates but also adenovirus and even rhinovirus isolates (data not shown). The cell types chosen are not optimal for the culture of rhinovirus, but that was not of primary interest in our investigation. In addition, influenza virus, RSV, and adenoviruses were detected in nasopharyngeal aspirates by commercial antigen detection assays.
The rather strict separation of the PIV1 outbreak and the influenza A outbreak, with one virus completely replacing the other one in the community with almost no overlap, was an interesting observation. In contrast, RSV was also detected throughout the influenza A period, yet in the particular year studied, no pronounced RSV epidemic was observed in our patients. This is probably due to the low birth rate, which results in sufficient numbers of susceptible individuals necessary for efficient transmission kinetics only every 2–3 years. In contrast, in countries with higher birth rates, strong RSV outbreaks seem to occur on an annual basis, with numbers of cases far exceeding the hMPV cases [12]. Furthermore, the relatively low overall population density and rural character of our region may play a role, since increases in the incidence of RSV and influenza A virus have been correlated with urbanisation in long-term epidemiological surveys [13].
During the peak season of influenza A, there were no isolations of hMPV in our paediatric study group. The very few isolations of hMPV in this time period were obtained in our laboratory from adult lung transplant patients only. In these immunocompromised patients, prolonged persistence of hMPV is observed, and a different virus subtype has even been sequenced in the interepidemic time period [14]. The latter finding could be an indication for independently transmitted hMPV strains within the hospital environment (nosocomial infections?), which, in adult transplant patients, may lead to rather severe sequelae [14].
The coincidence of the community outbreak of PIV1 with the peak incidence of hMPV was obvious, a situation that was also observed in the following year in the nearby South Tyrolean region (unpublished observation). Whether this might indicate similar physico/chemical properties of the two viruses and/or comparable means of transmission remains to be clarified. As most of the dual infections observed were a combination of PIV1 and hMPV, one could also speculate that a possible synergy between these viruses might exist, leading to cocirculation in the community. Some investigators, who have reported the frequent detection of hMPV in combination with the SARS virus, have recently proposed such a concept [8]. hMPV coinfection was suspected to enhance the early transmission of the less contagious coronavirus, thus contributing to the development of so-called “superspreaders” [15].
It is not clear at present whether dual infections contribute to the severity of symptoms and the rate of hospitalisation. In a recent multicentre study in Germany, König et al. [5] reported that only 1% of all hMPV infections were observed in children who did not require ICU support, yet among children in the ICU, hMPV was detected in 30%, with an impressive 60% of hMPV infections being associated with RSV infection. The relatively low number of RSV dual infections in our study could also be due to the reduced sensitivity of the commercial RSV antigen detection test as compared to PCR methods, but the data are difficult to compare because the König study, in contrast to ours, did not target actively replicating virus but instead detected nucleic acid material in nasopharyngeal samples directly. Thus, the amplification of residual genetic material from previous infections remains an obstacle when interpreting the results of highly sensitive PCR methods [1, 2]. Since the majority of hMPV isolations in our study were not coinfections with other viruses and the overall number of hospitalised children in whom hMPV was the only virus detected was relatively high (14%), we do not have any reason to assume that this particular hMPV strain is less virulent than RSV. However, recent data suggests that, in Italy, RSV is associated with higher hospitalisation rates than hMPV [3]. In addition, a significantly longer duration of symptoms has been reported for RSV compared with hMPV [4].
Only a few respiratory adenoviruses (type 2) were detected with both PCR and the antigen detection assays, which leads us to conclude that children with respiratory adenovirus strains are rarely hospitalised in our population.
Mimiviruses were not detected at all in our study, which, retrospectively, is not too surprising. According to the most recent publication by LaScola et al. [7], who discovered the mimiviruses, these viruses seem to be found only in severely ill ICU patients, which does not describe most of our paediatric study group. Since mimiviruses also were not detected in several samples of amoebae-containing water obtained from water tanks, one can assume that this particular virus subtype is either not highly abundant or is subject to regional variances in prevalence that have not yet been studied. In conclusion, this first investigation of Mimiviridae in paediatric patients demonstrates that, at present, there is no indication that mimiviruses contribute significantly to paediatric respiratory illness in Austria.
Acknowledgements
The authors are most grateful to Didier Raoult (Marseille, France) for generously providing his mimivirus “suicide PCR” protocol at a very early stage. Samples containing amoebae originating from water tanks in Austria and Thailand were kindly provided by F. Tiefenbrunner (Innsbruck, Austria). Thanks also to Barbara Huck (Freiburg, Germany) and Theresia Popow-Kraupp (Vienna, Austria) for providing controls for hMPV and parainfluenza testing, respectively.
References
- 1.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 2.Schwarze J, O’Donnell DR, Rohwedder A, Openshaw PJ. Latency and persistence of respiratory syncytial virus despite T cell immunity. Am J Respir Crit Care Med. 2004;169:801–805. doi: 10.1164/rccm.200308-1203OC. [DOI] [PubMed] [Google Scholar]
- 3.Bosis S, Esposito S, Niesters HG, Crovari P, Osterhaus AD, Principi N. Impact of human metapneumovirus in childhood: comparison with respiratory syncytial virus and influenza viruses. J Med Virol. 2005;75:101–104. doi: 10.1002/jmv.20243. [DOI] [PubMed] [Google Scholar]
- 4.Viazov S, Ratjen F, Scheidhauer R, Fiedler M, Roggendorf M. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J Clin Microbiol. 2003;41:3043–3045. doi: 10.1128/JCM.41.7.3043-3045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.König B, König W, Arnold R, Werchau H, Ihorst G, Forster J. Prospective study of human metapneumovirus infection in children less than 3 years of age. J Clin Microbiol. 2004;42:4632–4635. doi: 10.1128/JCM.42.10.4632-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkesmann A, Schildgen O, Eis-Hubinger AM, Geikowski T, Glatzel T, Lentze MJ, Bode U, Simon A. Human metapneumovirus infections cause similar symptoms and clinical severity as respiratory syncytial virus infections. Eur J Pediatr. 2006;165:467–475. doi: 10.1007/s00431-006-0105-4. [DOI] [PubMed] [Google Scholar]
- 7.LaScola B, Marrie TJ, Auffray JP, Raoult D. Mimivirus in pneumonia patients. Emerg Infect Dis. 2005;11:449–452. doi: 10.3201/eid1103.040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan PK, Tam JS, Lam CW, Chan E, Wu A, Li CK, Buckley TA, Ng KC, Joynt GM, Cheng FW, To KF, Lee N, Hui DS, Cheung JL, Chu I, Liu E, Chung SS, Sung JJ. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis. 2003;9:1058–1063. doi: 10.3201/eid0909.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gröndahl B, Puppe W, Hoppe A, Kuhne I, Weigl JA, Schmitt HJ. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maertzdorf J, Wang CK, Brown JB, Quinto JD, Chu M, de Graaf M, van den Hoogen BG, Spaete R, Osterhaus AD, Fouchier RA. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–986. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, Drancourt M. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. Proc Natl Acad Sci USA. 2000;97:12800–12803. doi: 10.1073/pnas.220225197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noyola DE, Alpuche-Solis AG, Herrera-Diaz A, Soria-Guerra RE, Sanchez-Alvarado J, Lopez-Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. J Med Microbiol. 2005;54:969–974. doi: 10.1099/jmm.0.46052-0. [DOI] [PubMed] [Google Scholar]
- 13.Glezen WP. The changing epidemiology of respiratory syncytial virus and influenza: impetus for new control measures. Pediatr Infect Dis J. 2004;23(Suppl 11):S202–S206. doi: 10.1097/01.inf.0000144662.86396.07. [DOI] [PubMed] [Google Scholar]
- 14.Larcher C, Geltner C, Fischer H, Nachbaur D, Müller LC, Huemer HP. Human metapneumovirus infection in lung transplant recipients: clinical presentation and epidemiology. J Heart Lung Transplant. 2005;24:1891–1901. doi: 10.1016/j.healun.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Bassetti S, Bischoff WE, Sherertz RJ. Are SARS superspreaders cloud adults? Emerg Infect Dis. 2005;11:637–638. doi: 10.3201/eid1104.040639. [DOI] [PMC free article] [PubMed] [Google Scholar]


