Abstract
We have developed an aptameric enzyme subunit (AES) which can detect the DNA in a homogeneous solution. The AES is an artificial enzyme subunit composed of an enzyme-inhibiting aptamer bearing a target-molecule binding site. We connected a probe DNA to a thrombin-inhibiting aptamer at its 5′ or 3′ end. The inhibitory activity of the thrombin-inhibiting aptamer bearing the probe DNA decreased compared to that of the original aptamer; however, it recovered upon hybridization with the target DNA. Using this AES, we were able to detect target DNAs by measuring the thrombin activity in a homogeneous solution.
Keywords: Aptameric enzyme subunit, Enzyme-inhibiting aptamer, Homogeneous DNA sensing
Introduction
A homogeneous detection, which does not require the bound/free separation, allows us to detect target molecules quickly and easily. Designed probe DNAs modified fluorescent dyes have been used mainly in homogeneous DNA sensing: molecular beacons (Tyagi et al. 1996), double-stranded probes (Li et al. 2002), excimer fluorescence (Masuko et al. 1998), light-up probe (Isacsson et al. 2000), fluorescent polymers (Ho et al. 2002), and DNAzymes (Wang et al. 2001; Stojanovic et al. 2001; Xiao et al. 2004; Tian et al. 2006). However, the use of enzymes can improve the sensitivity of detection since enzymes can amplify the detection signals through enzymatic reactions. At present, there are DNA sensing systems which use enzymes for homogeneous assays (Saghatelian et al. 2003; Pavlov et al. 2005); however, the inhibitors and probe DNAs must be covalently attached to the enzymes.
We developed an aptameric enzyme subunit (AES), which does not require any modification for use in homogeneous detection systems (Yoshida et al. 2006). The AES is an aptamer composed of an enzyme-inhibiting aptamer and a target-molecule-binding aptamer. Aptamers are nucleic acid ligands which can bind their target molecules with high affinity and specificity (Ellington et al. 1990; Tuerk et al. 1990). A target-molecule-binding aptamer was inserted into an enzyme-inhibiting aptamer to enable the change of the structure of an enzyme-inhibiting aptamer upon the recognition of the target molecule. Therefore the inhibitory activity of the AES was changed by binding to the target molecule, that is, we can detect the target molecule by measuring the corresponding enzymatic activity in a homogeneous solution.
We also applied the AES to a DNA sensing system by inserting a stem-and-loop structure into an enzyme-inhibiting aptamer (Yoshida et al. 2006). However, we have to consider the free energy of hybridization of the stem site and the loop site for the design of an AES bearing a stem-and-loop structure, since the stem site must fold into a double helix structure in the absence of the target DNA and be dehybridized by the hybridization of the target DNA with the loop site. Moreover, we also have to consider the free energy of the folding of the enzyme-inhibiting aptamer. It is not easy to construct AESs which can detect various target DNAs with this design.
Here, we present a novel AES design strategy (Fig. 1). This AES design is very simple, since the only requirement is for the AES to connect the probe DNA to end of the enzyme-inhibiting aptamer. Therefore we do not need to consider the secondary structure and the free energy of the folding of the enzyme-inhibiting aptamer like previous AES. We connected several kinds of probe DNA to end of the thrombin-inhibiting aptamer and investigated the changes in thrombin activity which resulted from the hybridization of these AESs with their target DNAs.
Fig. 1.
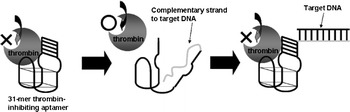
DNA sensing scheme based on an AES bearing a probe DNA at the end of the thrombin-inhibiting aptamer
Materials and methods
Materials
Human thrombin was purchased from Wako Chemicals, and human fibrinogen was purchased from Wako Chemicals and Sigma. Dimethyl sulfate and 2-mercaptoethanol were purchased from Tokyo Kasei Kogyo, ethidium bromide form Sigma, and tRNA from baker’s yeast from Roche Diagnostics. All oligonucleotides were obtained from Invitrogen. The other reagents were of analytical grade.
Design of the aptameric enzyme subunit
The DNA sequences used in this study are shown in Table 1. We designed a thrombin-inhibiting aptamer, to which we attached 36-, 27- or 18-mer complementary sequences of target DNAs at its 5′ or 3′ end. We used three different DNA sequences as the target DNAs: a part of the Salmonella invA gene (nucleotide positions 426 to 452) was used for the AES invA series (Galan et al. 1990), a part of the SARS coronavirus genomic antisense sequence (genome location 15348 to 15383) (Rota et al. 2003; Marra et al. 2003) for the AES SARS 1 series and a part of the SARS coronavirus genomic sequence (genome location 15561 to 15596) for the AES SARS 2 series. The 36-, 27- and 18-mer DNA sequences were used as the target DNAs, and these complementary strands were added to the thrombin-inhibiting aptamer at its 5′ or 3′ end.
Table 1.
The sequences of the oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| Thrombin aptamer | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES invA 5′36 | CTTCAAATCGGCATCAATACTCATCTGTTTACCGGGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES invA 5′27 | GGCATCAATACTCATCTGTTTACCGGGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES invA 5′18 | ACTCATCTGTTTACCGGGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES invA 3′36 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGCTTCAAATCGGCATCAATACTCATCTGTTTACCGGG |
| AES invA 3′27 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGGGCATCAATACTCATCTGTTTACCGGG |
| AES invA 3′18 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGACTCATCTGTTTACCGGG |
| AES SARS-1 5′36 | CTCTATAGAAATAGGGATGTTGATCATGAATTCGTGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-1 5′27 | AATAGGGATGTTGATCATGAATTCGTGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-1 5′18 | GTTGATCATGAATTCGTGCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-1 3′36 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGCTCTATAGAAATAGGGATGTTGATCATGAATTCGTG |
| AES SARS-1 3′27 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGAATAGGGATGTTGATCATGAATTCGTG |
| AES SARS-1 3′18 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGGTTGATCATGAATTCGTG |
| AES SARS-2 5′36 | GCCGCCACACATGACCATCTCACTTAATACTTGCGCCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-2 5′27 | GCCGCCACACATGACCATCTCACTTAACACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-2 5′18 | GCCGCCACACATGACCATCACTGGTAGGTTGGTGTGGTTGGGGCCAGTG |
| AES SARS-2 3′36 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGGCCGCCACACATGACCATCTCACTTAATACTTGCGC |
| AES SARS-2 3′27 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGGCCGCCACACATGACCATCTCACTTAA |
| AES SARS-2 3′18 | CACTGGTAGGTTGGTGTGGTTGGGGCCAGTGGCCGCCACACATGACCAT |
| 36-mer invA | CCCGGTAAACAGATGAGTATTGATGCCGATTTGAAG |
| 27-mer invA | CCCGGTAAACAGATGAGTATTGATGCC |
| 18-mer invA | CCCGGTAAACAGATGAGT |
| 36-mer SARS-1 | CACGAATTCATGATCAACATCCCTATTTCTATAGAG |
| 27-mer SARS-1 | CACGAATTCATGATCAACATCCCTATT |
| 18-mer SARS-1 | CACGAATTCATGATCAAC |
| 36-mer SARS-2 | GCGCAAGTATTAAGTGAGATGGTCATGTGTGGCGGC |
| 27-mer SARS-2 | TTAAGTGAGATGGTCATGTGTGGCGGC |
| 18-mer SARS-2 | ATGGTCATGTGTGGCGGC |
| 27-mer control DNA | GAATCCGGTACTGGCTAAGACAACTGT |
| 27-mer Complementary DNA | GGCATCAAT ACTCATCTGTTTACCGGG |
The thrombin-inhibiting aptamer sequences are underlined
For the experiment involving the native-PAGE and DMS footprinting of AES invA 3′27, the AES invA 3′27, the 27-mer invA, the 27-mer control DNA and the thrombin-inhibiting aptamer were modified with fluorescein isothiocyanate (FITC) at their 5′ ends. For the measurement of the CD spectra of AES invA 3′27, the 27-mer complementary strand to the invA gene (27-mer Complementary DNA) which was added to the thrombin-inhibiting aptamer part of AES invA 3′27 was synthesized.
Measurement of the inhibitory effect of the aptamers on thrombin
Fifty μl of folding buffer (50 mM Tris/HCl, 5 mM KCl, pH 8.0) containing 4 μM aptamer and 4 μM target DNA or control DNA (final concentration 1 μM) were heat-denatured at 95°C for 3 min and cooled to 25°C over a period of 30 min. After the heat treatment, the AES solution was mixed with 50 μl of measurement buffer (50 mM Tris/HCl, 5 mM KCl, 100 mM NaCl, pH 8.0) containing thrombin at 54 nM, and the mixture was incubated for 5 min at 37°C. After 5 min, 100 μl AES solution containing thrombin were added to 100 μl measurement buffer containing fibrinogen at 1 mg/ml. The clotting time was measured using an automated fibrometer (Amelung KC-4A Micro Coagulation Analyzer) at 37°C. In the experiment on the influence of the target DNA concentration on AES invA 3′27, we used 30 nM AES invA 3′27, 13.5 nM thrombin and 1 mg fibrinogen/ml, which was purchased from Sigma.
Native PAGE of the oligonucleotides
FITC-labeled 27-mer invA (8.3 μM) and AES invA 3′27 (8.3 μM) were mixed with the folding buffer, and the mixture was heat-treated as described above. After the heat treatment, 6 μl of the mixture were electrophoresed on 11% polyacrylamide gel in 1 × TBE buffer (89 mM Tris, 89 mM H3BO3, 2 mM EDTA) at room temperature. FITC fluorescence imaging was carried out on a Typhoon 8600 (Amersham Biosciences), and the oligonucleotides were also stained with ethidium bromide.
CD spectrum measurement
The CD spectra of the thrombin-inhibiting aptamer, AES invA 3′27, AES invA 3′27 hybridized with the 27-mer invA, the 27-mer complementary strand to the invA gene and the 27-mer complementary strand to the invA gene hybridized with the 27-mer invA were measured. All oligonucleotides, at 25 μM, were heat-treated in the folding buffer as described above, and then 100 μl of the AES solution were added to 300 μl of the measurement buffer. The CD spectra were measured with a J-720 spectropolarimeter at 200∼320 nm using a 0.1-cm path-length cuvette at 25°C.
DMS footprinting
FITC-labeled AES invA 3′27 (10 μM) was heat-treated as described above in the presence or absence of 10 μM 27-mer invA or 27-mer control DNA. Dimethyl sulfate (DMS) was added to give 0.1% (v/v) in a total volume of 100 μl, and the solutions were incubated at room temperature for 10 min. The methylation reaction was halted by adding 5 μl stop solution (1.5 M sodium acetate pH 7.0, 1 M 2-mercaptoethanol and 100 μg tRNA/ml). The reactants were precipitated using ethanol, dissolved in 100 μl 10% (w/v) piperidine, and incubated at 90°C for 30 min. After the piperidine cleavage, the sample solution was lyophilized and separated on 7 M urea/20% polyacrylamide gel, and FITC fluorescence imaging was carried out on a Typhoon 8600 (Amersham Biosciences).
Results and discussion
AES inhibitory activity change induced by the hybridization with the target DNA
We used a 31-mer thrombin-inhibiting aptamer which was screened by means of an evolution-mimicking algorithm using a library that contains a 15-mer prototype thrombin-inhibiting aptamer (Ikebukuro et al. 2005). Therefore, the 31-mer thrombin-inhibiting aptamer contained the consensus sequence of the 15-mer thrombin-inhibiting aptamer that had already been determined to fold into a G-quartet structure (Bock et al. 1992; Macaya et al. 1993; Schultze et al. 1994; Kuryavyi et al. 2001). Thus, the 31-mer thrombin-inhibiting aptamer used in this study was expected to form a G-quartet structure.
The 15-mer thrombin-inhibiting aptamer should not inhibit thrombin in the absence of potassium ions, since it must fold into a G-quartet structure. The 31-mer also lost its inhibitory activity for thrombin in the absence of potassium ions (data not shown). These results indicate that the structure of the thrombin-inhibiting aptamer is crucial for thrombin inhibition.
The DNA probe might partially hybridize with the thrombin-inhibiting aptamer site of the AES, thus possibly hindering the folding of the thrombin-inhibiting aptamer. Hybridization of the target DNA to the probe DNA site of AES would reduce the hybridization between the thrombin-inhibiting aptamer site and the connected probe DNA site, thus restoring the thrombin-inhibiting aptamer’s inhibitory activity. We therefore expected to be able to detect the target DNA using the designed AES and measuring the thrombin activity.
First, we constructed an AES invA series containing a 36-, 27- or 18-mer complementary strand of a part of the Salmonella invA gene at the 5′ or 3′ end of the thrombin-inhibiting aptamer. Secondary structure predictions carried out using the mfold program (Zuker et al. 2003) yielded a number of possible structures for each AES invA, all of which may be present in solution. We classified the secondary AES structure into two states. One is the native thrombin-inhibiting aptamer structure which bears the connected probe DNA at its end; the other is a thrombin-inhibiting aptamer structure disrupted by the hybridization of the connected probe DNA with the thrombin-inhibiting aptamer site. AES invA 5′36, 5′27, 5′18 and 3′27 (AES invA 5′36 is the thrombin-inhibiting aptamer which bears the 36-mer probe DNA at the 5′ end; the other AESs were named in the same way) were predicted to fold into both of these two states. In contrast, AES invA 3′36 and 3′18 were predicted to fold into only one state, the native thrombin-inhibiting aptamer structure. We assumed that the thrombin-inhibitory activity of AES invA 5′36, 5′27, 5′18 and 3′27 would decrease since the connected probe DNA would prevent the AESs from folding into the native thrombin-inhibiting aptamer structure, and that their inhibitory activity would recover upon the hybridization of their target DNAs with the connected probe DNA site since the prevention of folding into the native thrombin-inhibiting aptamer structure would be canceled. In contrast, the inhibitory activity of AES invA 3′36 and 3′18 would not decrease, since the probe DNAs would not prevent the AESs from folding into the native thrombin-inhibiting aptamer structure.
The thrombin-inhibiting activities of the AESs were determined by measuring the coagulation activity of thrombin in their presence. In the presence of thrombin only, the clotting time was approximately 10 s (Fig. 2A, lane 1); the addition of the thrombin-inhibiting aptamer led to the extension of the clotting time to approximately 110 s, as it inhibited thrombin activity (lane 2). The clotting times were approximately 75, 68, 86, and 52 s in the presence of AES invA 5′36, 5′27, 5′18 and 3′27, respectively (lanes 3, 5, 7 and 11); however, they increased to 129, 124, 119 and 113 s, respectively (lanes 4, 6, 8 and 12), upon the addition of the target DNAs. These results indicate that they have a lower thrombin-inhibitory capacity than the original thrombin-inhibiting aptamer; however, their lost inhibitory capacity can be recovered by the addition of their target DNAs. AES invA 5′36, 5′27, 5′18 and 3′27 did not lose their inhibitory activity completely. These results show that there was an equilibrium between the native thrombin-inhibiting aptamer structure and some of the disrupted thrombin-inhibiting aptamer structures, and that the equilibrium shifted to the native thrombin-inhibiting aptamer state upon hybridization with the target DNAs.
Fig. 2.
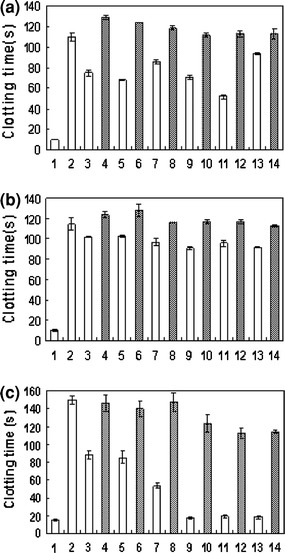
Inhibition of fibrinogen clotting by the addition of an AES invA (A), AES SARS-1 (B) or AES SARS-2 (C) series. These experiments were performed using 1 μM aptamer, 1 μM target DNA, 54 nM thrombin and 1 mg/ml fibrinogen at 37°C. Lane 1, thrombin only; lane 2, thrombin-inhibiting aptamer; lanes 3 and 4, AES which has a 36-mer probe DNA at its 5′ end; lanes 5 and 6, AES which has a 27-mer probe DNA at its 5′ end; lanes 7 and 8, AES which has an 18-mer probe DNA at its 5′ end; lanes 9 and 10, AES which has a 36-mer probe DNA at its 3′ end; lanes 11 and 12, AES which has a 27-mer probe DNA at its 3′ end; lanes 13 and 14, AES which has an 18-mer probe DNA at its 3′ end. The black bars indicate the absence of the target DNA, and the white bars indicate the presence of the target DNA. The clotting times are given as averages of three experiments, and error bars are provided
AES invA 3′36 and 3′18 were not expected to lose their thrombin-inhibitory capacity, since they were predicted to only form the native thrombin-inhibiting aptamer structure. However, their thrombin-inhibitory activities were lower than that of the original thrombin-inhibiting aptamer, and their lost inhibitory activities recovered upon the addition of their target DNAs (Fig. 2A, lanes 9, 10, 13 and 14). The inhibitory activities of AES invA 3′36 and 3′18 in the absence of their target DNAs were higher than that of AES invA 3′27. These results indicate that AES invA 3′36 and 3′18 fold into both native and disrupted thrombin-inhibiting aptamer structures but have a higher level of native structure than AES invA 3′27. This implies that this DNA sensing system can be effectively applied to target DNAs whose probe DNAs do not hybridize with the part of the AES containing the thrombin-inhibiting aptamer site. Therefore, we expect that this DNA sensing system can be applied to many DNA sequences.
We constructed an AES SARS 1 series containing the 36, 27 or 18-mer complementary strand of the SARS coronavirus genomic antisense sequences at the 5′ or 3′ end of the thrombin-inhibiting aptamer, to investigate whether this system is applicable to many kinds of target DNA sequences. These connected probe DNAs were predicted not to hybridize with the part of the AES containing the thrombin-inhibiting aptamer site. Their thrombin-inhibitory activities decreased compared to that of the thrombin-inhibiting aptamer, and their lost inhibitory activities recovered upon the addition of their target DNAs (Fig. 2B). We also constructed other AESs which have the 27-mer complementary strand of the influenza A virus membrane protein 1 gene sequence (nucleotide positions 150 to 176) at the 5′- or 3′- end of the thrombin-inhibiting aptamer (Fouchier et al. 2000). The mfold program predicted that the connected probe DNA would not hybridize with the thrombin-inhibiting aptamer site either. These AESs had the same characteristics as the AES SARS 1 series; that is, their inhibitory effect on thrombin decreased, recovering when their target DNAs were added (data not shown). These results indicate that this DNA sensing system can be applied to many DNA sequences.
We finally constructed an AES SARS-2 series containing the 36-, 27- or 18-mer complementary strand of a part of the SARS coronavirus genomic sequence at the 5′- or 3′- end of the thrombin-inhibiting aptamer. We presumed that if we selected probe DNAs that can tightly hybridize with the thrombin-inhibiting aptamer site, the AES would lose its thrombin-inhibitory capacity completely. Thus, when their target DNAs hybridize with probe DNA site and the thrombin-inhibitory capacity is recovered, the target DNAs would be detected with greater sensitivity than by using other AESs. The mfold program predicted that although AES SARS-2 5′36, 5′27 and 5′18 would fold into both the native thrombin-inhibiting aptamer structure and disrupted thrombin-inhibiting aptamer structures, AES SARS-2 3′36, 3′27 and 3′18 would fold only into disrupted thrombin-inhibiting aptamer structures. Therefore, we assumed that the former would show some inhibitory activity but not the latter.
The clotting times were approximately 17, 19 and 18 s in the presence of AES SARS-2 3′36, 3′27 and 3′18, respectively (Fig. 2C, lanes 9, 11 and 13), although the clotting times were approximately 88, 85 and 53 s in the presence of AES SARS-2 5′36, 5′27 and 5′18, respectively (lanes 3, 5 and 7). The inhibitory activity of AES SARS-2 3′36, 3′27 and 3′18 increased upon the addition of their target DNAs to 124, 113 and 114 s, respectively (lanes 10, 12 and 14). These results indicate that AES SARS-2 3′36, 3′27 and 3′18 lost almost all of their thrombin-inhibitory capacity; however, their target DNAs were able to induce a recovery of their inhibitory activities. Therefore, we can say that the target DNAs were detected with more sensitivity using the designed AESs than using other AESs.
Figure 2A, lane 2 and Fig. 2C, lane 2 show the same thrombin-inhibitory activity in the 31-mer thrombin-inhibiting aptamer; however, there is a time difference of 30 s. These experiments were performed under the same conditions, but the thrombin, fibrinogen and aptamer lots were different. This caused the discrepancies between Fig. 2A and 2C. However, when we used the same reagent lots, the discrepancies became very slight, as indicated by the error bars in Fig. 2.
Hybridization of AES invA 3′27 with its target DNA
The hybridization of AES invA 3′27 with its target DNA was confirmed by native PAGE on 11% polyacrylamide gel using FITC-labeled target DNA. The oligonucleotides were detected by both ethidium bromide staining and FITC fluorescence. In the presence of AES invA 3′27, while the band of the 27-mer control DNA did not shift (Fig. 3, lane 3), the band of the target DNA shifted to the AES invA 3′27 band (lane 4). This result indicates that AES invA 3′27 hybridized with its target DNA.
Fig. 3.
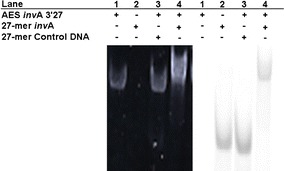
Native PAGE of the DNA complexes. The oligonucleotides (50 pmol) were electrophoresed on 11% polyacrylamide gel in 1 × TBE buffer at room temperature. The DNA was detected by ethidium bromide (left) and fluorescence (right). Both figures show the same gel. Lane 1, AES invA 3′27; lane 2, FITC-labeled 27-mer invA; lane 3, AES invA 3′27 with FITC-labeled 27-mer control DNA; lane 4, AES invA 3′27 with FITC-labeled 27-mer invA
Structural change in AES invA 3′27 upon hybridization with its target DNA
We expected that the thrombin-inhibiting aptamer would form a G-quartet structure, which would be important for the inhibitory activity of the AES complex. Therefore, we investigated whether or not a change in the G-quartet structure of the AES invA 3′27 occurs upon hybridization with the target DNA. We measured the CD spectra of AES invA 3′27 in the presence or absence of its target DNA and analyzed them through DMS footprinting assay.
CD spectra
The CD spectra were measured of the thrombin-inhibiting aptamer, AES invA 3′27, AES invA 3′27 hybridized with the 27-mer invA, the complementary strand of the 27-mer invA and the complementary strand of the 27-mer invA hybridized with the 27-mer invA. The CD spectrum of the thrombin-inhibiting aptamer showed three positive peaks, one near 295 nm, one near 250 nm and one near 210 nm, and two negative peaks, one near 265 nm and another near 235 nm (Fig. 4). This CD spectrum is almost identical to that of the 15-mer thrombin-inhibiting aptamer which folds into an anti-parallel G-quartet structure (Smirnov et al. 2000; Dapic et al. 2003), as previously determined by NMR and X-ray structural analyses. This suggests that the thrombin-inhibiting aptamer used in this study folds into a G-quartet structure.
Fig. 4.
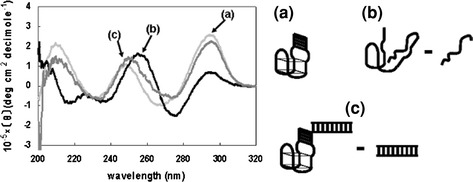
CD spectra of AES invA 3′27. Oligonucleotides at a concentration of 25 μM were used. Thrombin-inhibiting aptamer (a), CD spectrum of the probe DNA subtracted from that of AES invA 3′27 (b), CD spectrum of the probe DNA hybridized with the 27-mer invA subtracted from that of AES invA 3′27 hybridized with the 27-mer invA (c). The CD spectra were measured using a 0.1-cm path-length cuvette at 25°C
The CD spectrum produced by subtracting the spectrum of the complementary strand of the 27-mer invA from that of AES invA 3′27 showed peaks different from those of the thrombin-inhibiting aptamer; however, the CD spectrum produced by subtracting the spectrum of the complementary strand of the 27-mer invA hybridized with the 27-mer invA from that of AES invA 3′27 hybridized with the 27-mer invA showed peaks similar to the CD spectrum of the thrombin-inhibiting aptamer. These results suggest that the folding of AES invA 3′27 into a G-quartet structure is induced by the hybridization of the 27-mer invA with AES invA 3′27.
DMS footprinting
We also investigated, using the DMS footprinting assay, whether or not the G-quartet structure of AES invA 3′27 recovers upon hybridization with the target DNA. Dimethyl sulfate (DMS) methylates the N7 of guanine; however, an oligonucleotide folded into a G-quartet structure is protected from N7 methylation, since the N7 of one guanine forms a hydrogen bond with the N2 of another guanine to form a G-quartet structure. The DMS footprinting assay was performed on FITC-labeled AES invA 3′27, FITC-labeled AES invA 3′27 hybridized with its target DNA, and FITC-labeled thrombin-inhibiting aptamer in the presence or absence of potassium ions. In the thrombin-inhibiting aptamer, the potassium ions are necessary for G-quartet structure folding. Therefore, as shown Fig. 5, the guanine residues composing the G-quartet structure (G9, G10, G13, G14, G18, G19, G22 and G23) of the thrombin-inhibiting aptamer were exposed to N7 methylation in the absence of potassium ions (lanes 1 and 2). In AES invA 3′27, the guanine residues composing the G-quartet structure were exposed to N7 methylation in the presence or absence of potassium ions (lanes 3 and 4). However, these residues were protected from N7 methylation by the hybridization with the target DNA, although protection from N7 methylation was not detected when control DNA was added (lanes 5 and 6). These results also indicate that the folding of AES invA 3′27 into a G-quartet structure is induced by the hybridization of the 27-mer invA with AES invA 3′27.
Fig. 5.
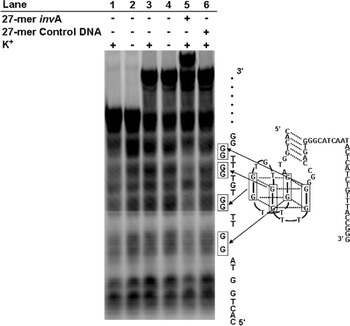
DMS footprinting assays of AES invA 3′27. Lane 1, thrombin-inhibiting aptamer with K+; lane 2, thrombin-inhibiting aptamer without K+; lane 3, AES invA 3′27 with K+; lane 4, AES invA 3′27 without K+; lane 5, AES invA 3′27 hybridized with the 27-mer invA with K+; lane 6, AES invA 3′27 containing the 27-mer control DNA with K+. The methylated guanines in the G-quartet structure are shown in the presumed secondary structure of AES invA 3′27. The experimental conditions are described in the “Materials and Methods” section
Calibration of the target DNA using AES invA 3′27
We obtained a calibration graph using 13.5 nM thrombin, 1 mg fibrinogen/ml and 30 nM AES invA 3′27. Under these conditions, the clotting time was approximately 36 s in the presence of thrombin only (data not shown). The addition of 30 nM AES invA 3′27 led to the extension of the clotting time to approximately 54 s. An increase in the clotting time from 54 s to 84 s was observed with the increase in the concentration of the target DNA to 30 nM, and when target DNA at concentrations higher than 30 nM was added, the clotting time became saturated (Fig. 6). The thrombin-inhibiting activity was not influenced by the addition of the control DNA. This calibration graph indicates that the target DNA of AES invA 3′27 was selectively detected by measuring the thrombin enzymatic activity in a homogeneous solution, and that the lower limit of target DNA detection was 10 nM.
Fig. 6.
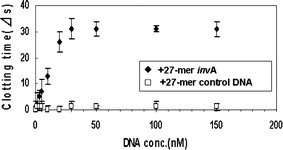
Calibration graph for the 27-mer invA target DNA using the AES invA 3′27. These experiments were performed using 30 nM AES 3′27 invA, 13.5 nM thrombin and 1 mg/ml fibrinogen. The clotting times are given as averages of three experiments, and error bars are provided
When we used 1 μM AES invA 3′27, the dynamic range extended from 0.1 μM to 1 μM (data not shown). The clotting time was saturated when AES invA 3′27 and its target DNA reached the same concentration. This result indicates that AESs bind to their target DNAs at a ratio of one to one, and that the dynamic range can be changed by changing the concentration of the AESs.
AES has some important advantages over other homogeneous DNA detection methods.
Because AES uses an enzyme which can amplify the detection signals, highly sensitive detection can be expected compared with FRET or DNAzyme-based technics. In this study, we used a thrombin-inhibiting aptamer; however, the sensitivity of the sensing system can be improved if a more suitable enzyme-inhibiting aptamer is developed for this sensing system.
AES does not require any modification. Although Saghatelian et al. (2003) and Pavlov et al. (2005) developed DNA sensing methods that use enzyme to detect the target DNA, these systems need to connect the probe DNA to the enzyme and the enzyme inhibitor. Enzyme modification usually results in significant loss of activity and requires a cumbersome process of enzyme preparation. In contrast, AES can be constructed very easily since AES is simply an oligonucleotide.
Detection of genome DNA may involve the amplification of several tens of sequences by PCR followed by detection by AES because non-specific products are frequently amplified during PCR. Simple detection of PCR products is very important, especially for the detection of such things as pathogenic bacteria or viruses. Therefore the combination of PCR and AES will be a powerful tool for the detection of pathogenic bacteria or viruses specifically.
Acknowledgements
This work was supported by Industrial Technology Research Grant Program in 2006 from New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
K. Ikebukuro and W. Yoshida have contributed equally to this work.
References
- Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HA, Boissinot M, Bergeron MG, Corbeil G, Dore K, Boudreau D, Leclerc M (2002) Angew. Chem., Int. Ed. Colorimetric and Fluorometric Detection of Nucleic Acids Using Cationic Polythiophene Derivatives. 114:1618–1621 [DOI] [PubMed]
- Ikebukuro K, Okumura Y, Sumikura K, Karube I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005;33:e108. doi: 10.1093/nar/gni108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isacsson J, Cao H, Ohlsson L, Nordgren S, Svanvik N, Westman G, Kubista M, Sjoback R, Sehlstedt U. Rapid and specific detection of PCR products using light-up probes. Mol Cell Probes. 2000;14:321–328. doi: 10.1006/mcpr.2000.0321. [DOI] [PubMed] [Google Scholar]
- Kuryavyi V, Majumdar A, Shallop A, Chernichenko N, Skripkin E, Jones R, Patel DJ. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple. J Mol Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
- Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci U. S. A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra MA, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Masuko M, Ohtani H, Ebata K, Shimadzu A. Optimization of excimer-forming two-probe nucleic acid hybridization method with pyrene as a fluorophore. Nucleic Acids Res. 1998;26:5409–5416. doi: 10.1093/nar/26.23.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov V, Shlyahovsky B, Willner I. Fluorescence detection of DNA by the catalytic activation of an aptamer/thrombin complex. J Am Chem Soc. 2005;127:6522–6523. doi: 10.1021/ja050678k. [DOI] [PubMed] [Google Scholar]
- Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Guckian KM, Thayer DA, Ghadiri MR. DNA detection and signal amplification via an engineered allosteric enzyme. J Am Chem Soc. 2003;125:344–345. doi: 10.1021/ja027885u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J Mol Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- Stojanovic MN, de Prada P, Landry DW. Catalytic molecular beacons. Chembiochem. 2001;2:411–415. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tian Y, He Y, Mao C. Cascade signal amplification for DNA detection. Chembiochem. 2006;7:1862–1864. doi: 10.1002/cbic.200600336. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Wang DY, Sen D. A novel mode of regulation of an RNA-cleaving DNAzyme by effectors that bind to both enzyme and substrate. J Mol Biol. 2001;310:723–734. doi: 10.1006/jmbi.2001.4811. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Pavlov V, Niazov T, Dishon A, Kotler M, Willner I. Catalytic beacons for the detection of DNA and telomerase activity. J Am Chem Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- Yoshida W, Sode K, Ikebukuro K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal Chem. 2006;78:3296–3303. doi: 10.1021/ac060254o. [DOI] [PubMed] [Google Scholar]
- Yoshida W, Sode K, Ikebukuro K. Homogeneous DNA sensing using enzyme-inhibiting DNA aptamers. Biochem Biophys Res Commun. 2006;348:245–252. doi: 10.1016/j.bbrc.2006.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


