Abstract
Cryptosporidiosis of calves is caused by the enteroprotozoan Cryptosporidium spp. The disease results in intense diarrhea of calves associated with substantial economic losses in dairy farming worldwide. The aim of this study was to determine calf, herd, and within-herd Cryptosporidium prevalence and identify Cryptosporidium species and subtypes in calves with diarrhea in intensive dairy herds in central Argentina. A total of 1073 fecal samples were collected from 54 randomly selected dairy herds. Cryptosporidium-oocysts were isolated and concentrated from fecal samples using formol-ether and detected by light microscopy with the modified Ziehl-Neelsen technique. Overall prevalence of oocyst-excreting calves was found to be 25.5% (274/1073) (95% C.I. 22.9; 28.1%). Of the herds studied, 89% (48/54) included at least one infected calf, whereas within-herd prevalence ranged from the absence of infection to 57% (20/35). A highly significant association was found between the presence of diarrhea and C. parvum infection (χ2 = 55.89, p < 0.001). For species determination, genomic DNA isolated from oocyst-positive fecal samples was subjected to PCR-RFLP of the 18S rRNA gene resulting exclusively in Cryptosporidium parvum identification. C. parvum isolates of calves displaying diarrhea and high rate of excretion of oocysts were subtyped by PCR amplification and direct sequencing of the 60 kDa glycoprotein (GP60) gene. Altogether five GP60 subtypes, designated IIaA18G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and IIaA24G1R1 were identified. Interestingly, IIaA18G1R1 and IIaA20G1R1 were predominant in calves with diarrhea and high infection intensity. Notably, IIaA24G1R1 represents a novel, previously unrecognized C. parvum subtype. The subtype IIaA18G1R1, frequently found in this study, is strongly implicated in zoonotic transmission. These results suggest that calves might be an important source for human cryptosporidiosis in Argentina.
Electronic supplementary material
The online version of this article (10.1007/s00436-019-06366-y) contains supplementary material, which is available to authorized users.
Keywords: Cryptosporidium parvum, GP60 subtype, Dairy calves, Diarrhea, Prevalence, Zoonotic transmission, Argentina
Introduction
Cryptosporidium spp. is an apicomplexan enteropathogen of a wide range of vertebrate hosts including humans. Cryptosporidial infection is common in calves and could cause severe diarrhea, lethargy, anorexia, and dehydration leading to substantial economic losses mainly related to reductions in weight gain and the application of palliative and preventive treatments. Although calves with cryptosporidiosis may recover after 4–6 weeks (Olson et al. 2004), a mortality rate of 35.5% in younger animals than 30 days has also been reported (Singh et al. 2006).
High herd prevalence of Cryptosporidium spp. has been reported in dairies, with values ranging from 50.5 to 96% (Hamnes et al. 2006; Brook et al. 2008; Trotz-Williams et al. 2008; Silverlås et al. 2009; Tiranti et al. 2011; Smith et al. 2014; Delafosse et al. 2015; Al Mawly et al. 2015). It has been reported that cryptosporidiosis risk is higher in calves younger than 30 days old (Tiranti et al. 2011; Smith et al. 2014; Delafosse et al. 2015; Avendaño et al. 2018).
Mainly, four Cryptosporidium species have been reported in cattle, Cryptosporidium parvum, Cryptosporidium bovis, Cryptosporidium andersoni, and Cryptosporidium ryanae (Santín et al. 2008; Ryan et al. 2014). Among them, C. parvum and C. bovis are the most common species identified in pre-weaned calves. C. parvum can be transmitted to humans via the fecal/oral route, following direct or indirect contact with infected animals (Ryan et al. 2014; Kinross et al. 2015). Cattle are recognized as a major parasite reservoir and contributor of zoonotic infection, because C. parvum subtypes infecting humans have been isolated from cattle (Xiao and Fayer 2008). C. parvum is an important zoonotic pathogen responsible for serious human diarrhea outbreaks in industrialized countries, affecting especially immunocompromised individuals (HIV patients), children, and the elderly (Xiao and Feng 2008). In developing countries, Cryptosporidium infection has been found to be associated with a significantly increased risk of deaths in toddlers with diarrhea aged 12–23 months (Kotloff et al. 2013).
Molecular tools have allowed to define and assess the genetic diversity of Cryptosporidium facilitating the quest to unravel transmission patterns and associated impacts on public health (Thompson and Ash 2016). In order to assess a possible transmission source of C. parvum, intraspecific subtyping based on the 60 kDa glycoprotein (GP60) gene is commonly used (Xiao 2010). Accordingly, identification of Cryptosporidium intraspecific variation has been central in understanding transmission dynamics of zoonotic parasite strains. Molecular characterization of C. parvum at the subtype level in dairy calves, important to determine zoonotic potential, have been conducted in several countries (Santín et al. 2008; Xiao and Fayer 2008; Amer et al. 2010; Silverlås et al. 2013; Smith et al. 2014; Kaupke and Rzeżutka 2015). In Argentina, two previous studies have reported C. parvum as the only species present in 120 pre-weaned dairy calves and subtypes IIaA16G1R1 IIaA17G1R1, IIaA18G1R1, IIaA19G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and IIaA23G1R1 have been identified (Tomazic et al. 2013; Del Coco et al. 2014). Whereas an association between subtypes and different clinical manifestations has been demonstrated in humans, few studies have tested the hypothesis that C. parvum-subtypes are associated with diarrhea intensity in dairy calves (Cama et al. 2007; Insulander et al. 2013; Adamu et al. 2014).
The aim of this study was to determine (i) Cryptosporidium prevalence in calves, herds, and within-herd and (ii) the presence of Cryptosporidium species and C. parvum subtypes in calves with diarrhea from intensive dairy farms in central Argentina.
Materials and methods
Study area
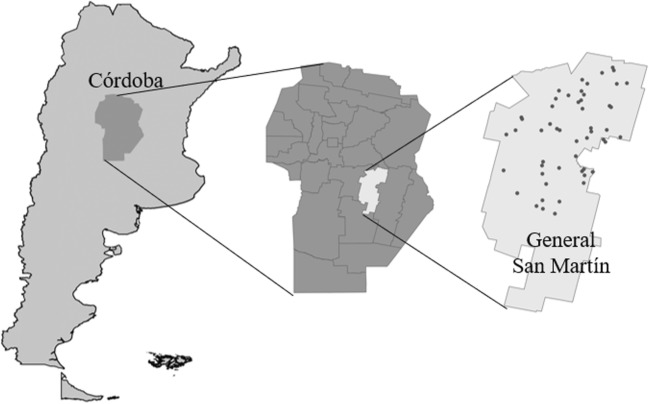
A cross-sectional study was carried out between April 2013 and March 2014 in the dairy area located in General San Martín district, Córdoba, Argentina, (Fig. 1, Supplementary Table 1). This area included 564 dairy herds (INDEC 2008), of which the herd size of 68% range from 101 to 500 cows (Sánchez et al. 2012).
Fig. 1.
Dairy herds sampled in General San Martín district, Córdoba, Argentina
Enrollment of farms
Farms were selected from a database of the Producer’s Rural Association, registered in the local execution unit Villa María, General San Martín district in Córdoba, Argentina. The inclusion criterion for dairy farms was a size of 101 to 300 cows as based on January 2013 data. Based on sample size calculations, a total of 60 farms were required to estimate a 19.3% prevalence with 9% precision and were randomly selected from 365 dairies (EPIDAT 3.1).
Sample collection
Exclusively calves younger than 60 days of age were sampled. Individual fecal samples were taken directly from the rectum using sterile plastic gloves and were stored at 4 to 8 °C until processing. At the moment of sample collection, feces consistency and presence of diarrhea were registered. Fecal consistency was scored as follows, solid (S), semisolid (SS), liquid (L), runny (R), and meconium (M). Diarrhea was defined as runny or liquid feces (Mcguirk 2008).
Laboratory methods
Samples were processed with the formol-ether concentration technique (Young et al. 1979); the supernatant was removed, leaving a final pellet volume of 2 ml. Fecal smears were prepared with 50 μl of each pellet. Presence of Cryptosporidium oocysts was detected by microscopic examination at × 1000 magnification using the modified Ziehl-Neelsen technique (Henriksen and Pohlenz 1981) (Fig. 2). Intensity of Cryptosporidium infection was scored semi-quantitatively according to the average number of oocysts in 20 randomly selected fields as follows: 0 (no oocysts), 1 (1–5 oocysts), 2 (6–10 oocysts), 3 (11–15 oocysts), and 4 (≥ 16 oocysts) (Chartier et al. 2013). All tests were performed by one person.
Fig. 2.
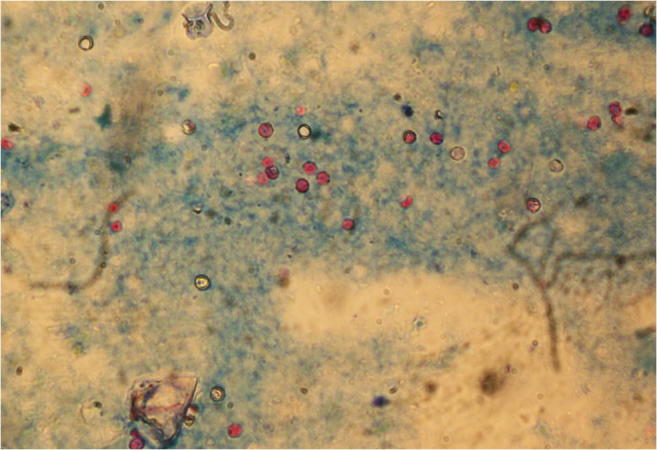
Cryptosporidium oocysts under microscopic examination of fecal smears at × 1000 magnification using the modified Ziehl-Neelsen technique
Typing and subtyping
Cryptosporidium oocysts were concentrated from oocyst-positive fecal samples using the sucrose flotation protocol of Coklin et al. (2007). Oocysts were ruptured with three freeze-thaw cycles, and total DNA was extracted with the QIAmp DNA Stool Mini Kit (Qiagen, Hilden, Germany) following the instructions of the manufacturer. For species determination, genomic DNA isolated from oocyst-positive fecal samples were subjected to PCR-RFLP as described in Xiao et al. (1999) including the use of an additional restriction enzyme MboII (Feng et al. 2007). For subtyping, C. parvum isolates were selected from calves with diarrhea and high infection intensity (scores 3 and 4), and the PCR amplicon of the gene encoding the 60 kDa glycoprotein (GP60) was directly sequenced and subtyped (Alves et al. 2003). GP60 subtypes were designated based on the number of trinucleotides TCA and TCG and the hexanucleotide ACATCA in the polymorphic tandem repeat region (Sulaiman et al. 2005). Nucleotide sequences were deposited in the GenBank database under accession numbers KX768771-KX768816.
Statistical analysis
Cryptosporidium parvum overall, herd, and within-herd prevalence was calculated for all calves. A chi-square test (χ2) was used to compare the frequency of C. parvum infection in diarrheic and healthy animals and to assess the association between calf age and the occurrence of C. parvum subtype-infection. All analyses were performed with the statistical software R 3.2.3.
Results
Descriptive data
From the 60 herds selected, six were excluded; since three farms had incorrect contact data, two farms send their calves to a collective artificial breeding system, and one farm had sold all animals at the time of the study. Altogether, a total of 1073 fecal samples were collected in the remaining 54 dairy herds corresponding to an average of 20 samples per herd (range 10 to 66). Of all sampled calves, 22% (236/1073) presented diarrhea (95% C.I. 19.5, 24.5).
Calves, herd, and within-herd Cryptosporidium parvum prevalence
Of 1073 sampled calves, 282 (26.3%) were positive for oocysts excretion after microscopic examination. Prevalence of oocyst excretion for calves of one, two, three, and four or more weeks of age was found to be 11.9% (12/101), 46.2% (141/305), 29.5% (99/336), and 9% (30/331), respectively.
Genomic DNA was isolated from oocyst-positive samples and subjected to PCR-RFLP for Cryptosporidium species determination. Eight samples with a low infection intensity (score 1) failed to amplify, whereas the PCR-RFLP pattern of the remaining 274 samples identified C. parvum resulting in a prevalence of 25.5% (95% C.I. 22.9; 28.1%) in the study area. Of the 274 C. parvum-positive samples, 35% (96) displayed a low (score 1), 41% (113) a medium (score 2), 13% (36) a high (score 3), and 11% (29) a very high (score 4) infection intensity. With regard of feces consistency, 17% (63/372) of C. parvum-positive calves displayed feces that had been graded to be of solid (S), 24% (104/432) semisolid (SS), 44% (39/89) liquid (L), 45% (66/147) runny (R), and 6% (2/33) meconium (M) consistency.
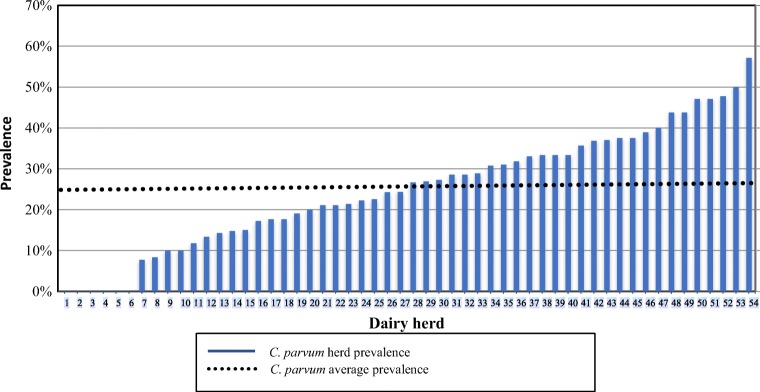
In 48 of 54 herds, at least one C. parvum positive calf was identified resulting in an overall herd prevalence of 89%. C. parvum within-herd prevalence was lower, ranging from 0 to 57% (mean 25%; first quartile 14.7%; third quartile 36%) and in 50% of herds, C. parvum within-herd prevalence was higher than average (Fig. 3). Diarrhea was absent in six herds; in three of these, no oocysts were detected, while in the three remaining only one or two C. parvum-positive calves with a low infection intensity (score 1) were found.
Fig. 3.
Cryptosporidium parvum prevalence by dairy herd (n = 54)
Cryptosporidium parvum subtyping
From 65 C. parvum-positive calves exhibiting a high (score 3) and very high infection intensity (score 4), 47 samples of diarrheal calves were selected for amplification and GP60 gene sequencing. All analyzed C. parvum GP60 subtypes belonged to the IIa family. Altogether, five C. parvum GP60 subtypes (IIaA18G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and IIaA24G1R1) could be identified (Table 1). Interestingly, in all GP60 alleles, a non-synonymous nucleotide exchange from “GAC” to “GGC” was observed, corresponding to an exchange of Asn to Gly at amino acid site 99, as previously reported (Tomazic et al. 2013).
Table 1.
Cryptosporidium parvum GP60 subtypes in dairy calves
| Subtype | Samples by age groups a | Total | Geographic region b | Reference | |||
|---|---|---|---|---|---|---|---|
| 1 week | 2 weeks | 3 weeks | 4 weeks | ||||
| IIaA18G1R1 | 0 | 5 | 6 | 1 | 12 | Argentina (1 of 46; 13 of 75) | (Tomazic et al. 2013, Del Coco et al. 2014) |
| England (2 of 51) | (Brook et al. 2009) | ||||||
| The Netherlands (2 of 129) | (Wielinga et al. 2008) | ||||||
| Sweden (3 of 171) | (Silverlås et al. 2013) | ||||||
| Czech Republic (3 of 137) | (Kváč et al. 2011) | ||||||
| Hungary (1 of 21) | (Plutzer and Karanis 2007) | ||||||
| Serbia and Montenegro (2 of 18) | (Misic and Abe 2007) | ||||||
| Brazil (1 of 28) | (do Couto et al. 2013) | ||||||
| IIaA20G1R1 | 1 | 16 | 7 | 1 | 25 | Argentina (8 of 46; 27 of 75) | (Tomazic et al. 2013, Del Coco et al. 2014) |
| Sweden (5 of 171) | (Silverlås et al. 2013) | ||||||
| Serbia and Montenegro (2 of 18) | (Misic and Abe 2007) | ||||||
| IIaA21G1R1 | 0 | 2 | 0 | 0 | 2 | Argentina (15 of 46; 10 of 75) | (Tomazic et al. 2013, Del Coco et al. 2014) |
| Sweden (11 of 171) | (Silverlås et al. 2013) | ||||||
| IIaA22G1R1 | 1 | 5 | 0 | 0 | 6 | Argentina (5 of 46; 16 of 75) | (Tomazic et al. 2013, Del Coco et al. 2014) |
| Sweden (7 of 171) | (Silverlås et al. 2013) | ||||||
| Czech Republic (12 of 137) | (Kváč et al. 2011) | ||||||
| IIaA24G1R1 | 0 | 2 | 0 | 0 | 2 | None | None |
| Total | 2 | 30 | 13 | 2 | 47 | ||
aFrequency of samples by age groups and C. parvum subtypes
bCountry in which the respective subtype has been reported in calves. Number of positive samples and the examined study group is given
A highly significant association was found between diarrhea presence and C. parvum infection (χ2 = 55.89, p < 0.001, Supplementary Table 2). The predominant subtype in calves with diarrhea and high infection intensity was IIaA20G1R1 (25/47), followed by IIaA18G1R1 (12/47). Additionally, a novel subtype, IIaA24G1R1 was determined in two calves.
No statistically significant relationship was found between calf age and a specific subtype presence (p = 0.49, Supplementary Table 3). Nevertheless, particular subtypes were present only in animals of a certain age. For example, IIaA21G1R1, IIaA22G1R1, and IIaA24G1R1 subtypes were observed only in animals of 2 weeks of age or less. In contrast, the IIaA18G1R1 subtype was found exclusively in calves 2 weeks of age or older, while the most predominant subtype, IIaA20G1R1, was identified in all ages. Furthermore, all subtypes were found to be not clustered but individually distributed in different dairy herds (Supplementary Table 4).
Discussion
Cryptosporidium parvum infection in calves is widespread (Brook et al. 2008; Trotz-Williams et al. 2008; Al Mawly et al. 2015; Qi et al. 2015), and also in the present study, C. parvum was, in agreement with previous reports in Argentina, the only species identified in calves (Tomazic et al. 2013; Del Coco et al. 2014). However, in contrast to previous studies done in Argentina, a considerably higher overall prevalence was found (Del Coco et al. 2008; Tiranti et al. 2011; Garro et al. 2016), probably due to an additional oocysts concentration step prior to microscopic examination significantly increasing the sensitivity of oocyst detection. In the present study, the sampling fraction of calves ≤ 30 days of age was 83% (888/1073), which is the age range were C. parvum is most frequently found (Santín et al. 2008; Xiao 2010; Ryan et al. 2014). Delafosse et al. (2015) found that a high infection intensity (scores 3 or 4) predicted an increased mortality rate of calves 90 days of age. Based on this finding, it is anticipated that 23.7% of oocyst-excreting calves (or 6.1% of the studied calves) in the present study may be subjected to an increased mortality.
At least one C. parvum-positive calf was detected in 89% of the herds studied. Highly variable herd prevalence ranging from 50.5 to 96% have been reported (Hamnes et al. 2006; Brook et al. 2008; Trotz-Williams et al. 2008; Silverlås et al. 2009; Tiranti et al. 2011; Smith et al. 2014; Al Mawly et al. 2015; Delafosse et al. 2015; Garro et al. 2016; Avendaño et al. 2018). Similar to the considerable differences of overall prevalence that have been reported, corresponding dissimilarities in herd prevalence may likewise be attributed to the different sensitivities of applied diagnostic test. Accordingly, Garro et al. (2016) estimated a herd prevalence of 67% (overall prevalence 16.3%) compared with 89% (overall prevalence 26.3%) in the present study. Other factors that may bias towards lower herd prevalence values, maybe the sampling of a smaller age range or a higher number of older animals (30–60 days). In addition, also, a lower number of calves sampled in each herd may significantly reduce the probability of finding positive calves particularly in herds with very low prevalence. Since only one fecal sample from each calf was examined, the true prevalence value may have been underestimated in the present study.
Diarrhea was not observed in 6 herds, 3 of which tested negative for all animals. Interestingly, the 3 remaining herds presented 1 or 2 C. parvum positive calves with low infection intensity score (score 1). A highly significant association was found between the presence of diarrhea and C. parvum infection, reinforcing the notion that this enteropathogen is most relevant for diarrhea development in neonatal dairy calves (Izzo et al. 2011; Al Mawly et al. 2015; Meganck et al. 2015; Garro et al. 2017). Nevertheless, the association of C. parvum with diarrhea may be overestimated since possible co-infections with other microorganisms were not evaluated.
Noteworthy, a novel subtype IIaA24G1R1 could be identified in two different calves of the same herd. Both animals showed profuse diarrhea, high-level of dehydration and, though treated, eventually died. Further laboratory testing excluded Salmonella spp., enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), Rotavirus, and Coronavirus as primary cause of death (data not shown).
Besides the novel IIaA24G1R1 subtype, the remaining 4 subtypes reported in this study have been previously detected in Argentina. High genetic variability has been found in dairy calves in Argentina. Tomazic et al. (2013) reported the six subtypes IIaA17G1R1, IIaA18G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and IIaA23G1R1 from 45 dairy calves. Del Coco et al. (2014) described the seven subtypes IIaA16G1R1, IIaA18G1R1, IIaA19G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and IIaA23G1R1 from 73 dairy calves. The predominant subtype IIaA20G1R1 detected in this study, was also found with relatively high frequency in dairy calves studied in other areas of Argentina (Tomazic et al. 2013; Del Coco et al. 2014), apart from Sweden (Silverlås et al. 2013), Serbia, and Montenegro (Misic and Abe 2007). The second most frequent IIaA18G1R1 subtype, seems to be more widely distributed, being reported in England (Brook et al. 2009), The Netherlands (Wielinga et al. 2008), Sweden (Silverlås et al. 2013), Czech Republic (Kváč et al. 2011), Hungary (Plutzer and Karanis 2007), Serbia and Montenegro (Misic and Abe 2007), Brazil (do Couto et al. 2013), and Argentina (Tomazic et al. 2013; Del Coco et al. 2014). In England, this subtype has been also identified in humans (Chalmers et al. 2011), strongly suggesting a zoonotic transmission risk. In contrast, a recent study carried out in Argentinean and Brazilian persons, C. hominis was frequently detected (Peralta et al. 2016) and none of C. parvum subtypes reported in this study were found.
Further research is required to understand the real clinical significance of each C. parvum subtype, as well as their interaction when co-infections occur. The presence of at least one C. parvum zoonotic subtype in dairy calves implies transmission to humans. Consequently, molecular epidemiology studies in humans, especially in farm workers that are in close contact with C. parvum-infected animals, may further substantiate the existence of zoonotic transmission of the parasite.
Conclusions
The current study revealed that Cryptosporidium parvum is widely distributed in dairy calf operations. The five C. parvum GP60 subtypes IIaA18G1R1, IIaA20G1R1, IIaA21G1R1, IIaA22G1R1, and a novel IIaA24G1R1 variant were identified. The most commonly detected subtypes in calves with diarrhea and high infection intensity score were IIaA20G1R1 and IIaA18G1R1. Subtype IIaA18G1R1 was frequently found in this study and is strongly implicated in zoonotic transmission suggesting that calves might be an important source of human cryptosporidiosis in Argentina. However, to confirm this hypothesis, studies in humans are required.
Electronic supplementary material
(DOCX 17 kb)
Funding
This project was supported by the Proyectos y Programas de Investigación-Universidad Nacional de Río Cuarto (Fomento No. A338), Agencia Nacional de la Promoción Científica y Tecnológica (ANPCyT) (PICT 2013-1708, PICT 2012-0695), and Fundación Universidad de Morón (PID 8-2015). The authors acknowledge the students Georgina Matteo, Santiago Hernandez, and Maria Luz Gomez Vega for sample processing and field visits and all participating producers.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics
All procedures performed in studies involving animals were in accordance with the standards of the Research Ethics Committee of the Universidad Nacional de Río Cuarto (approval number 47/11 and 173/16).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis. 2014;8(4):e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Mawly J, Grinberg A, Prattley D, Moffat J, Marshall J, French N. Risk factors for neonatal calf diarrhoea and enteropathogen shedding in New Zealand dairy farms. Vet J. 2015;203:155–160. doi: 10.1016/j.tvjl.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41:2744–2747. 10.1128/JCM.41.6.2744-2747.2003 [DOI] [PMC free article] [PubMed]
- Amer S, Honma H, Ikarashi M, Tada C, Fukuda Y, Suyama Y, Nakai Y. Cryptosporidium genotypes and subtypes in dairy calves in Egypt. Vet Parasitol. 2010;169:382–386. doi: 10.1016/j.vetpar.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Avendaño C, Ramo A, Vergara-Castiblanco C, Sánchez-Acedo C, Quílez J. Genetic uniqueness of Cryptosporidium parvum from dairy calves in Colombia. Parasitol Res. 2018;117:1317–1323. doi: 10.1007/s00436-018-5818-6. [DOI] [PubMed] [Google Scholar]
- Brook E, Hart CA, French N, Christley R. Prevalence and risk factors for Cryptosporidium spp. infection in young calves. Vet Parasitol. 2008;152:46–52. doi: 10.1016/j.vetpar.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Brook EJ, Anthony Hart C, French NP, Christley RM. Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Vet J. 2009;179:378–382. doi: 10.1016/j.tvjl.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Cama VA, Ross JM, Crawford S, Kawai S, Chavez-Valdez R, Vargas D, et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. JID. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- Chalmers RM, Smith RP, Hadfield SJ, Elwin K, Giles M. Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasit Res. 2011;108:1321–1325. doi: 10.1007/s00436-010-2199-x. [DOI] [PubMed] [Google Scholar]
- Chartier C, Rieux A, Delafosse A, Lehebel A, Paraud C. Detection of Cryptosporidium oocysts in fresh calf faeces: characteristics of two simple tests and evaluation of a semi-quantitative approach. Vet J. 2013;198:148–152. doi: 10.1016/j.tvjl.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Coklin T, Farber J, Parrington L, Dixon B. Prevalence and molecular characterization of Giardia duodenalis and Cryptosporidium spp. in dairy cattle in Ontario, Canada. Vet Parasitol. 2007;150:297–305. doi: 10.1016/j.vetpar.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Del Coco VF, Córdoba MA, Basualdo JA. Cryptosporidium infection in calves from a rural area of Buenos Aires. Argentina Vet Parasitol. 2008;158:31–35. doi: 10.1016/j.vetpar.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Del Coco VF, Córdoba MA, Bilbao G, Pinto de Almeida Castro A, Basualdo JA, Fayer R, et al. Cryptosporidium parvum GP60 subtypes in dairy cattle from Buenos Aires, Argentina. Res Vet Sci. 2014;96:311–314. doi: 10.1016/j.rvsc.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Delafosse A, Chartier C, Dupuya MC, Dumoulin M, Pors I, Paraud C. Cryptosporidium parvum infection and associated risk factors in dairy calves in western France. Prev Vet Med. 2015;118:406–412. doi: 10.1016/j.prevetmed.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Couto MCM, Lima MD, Bomfim TC. New Cryptosporidium parvum subtypes of IIa subfamily in dairy calves from Brazil. Acta Trop. 2013;130:117–122. doi: 10.1016/j.actatropica.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, Fayer R, Gatei W, Cama V, Xiao L. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Par. 2007;144:1–9. doi: 10.1016/j.vetpar.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Garro CJ, Morici GE, Utgés ME, Tomazic ML, Schnittger L. Prevalence and risk factors for shedding of Cryptosporidium spp. oocysts in dairy calves of Buenos Aires Province, Argentina. Parasite Epidemiol Control. 2016;1:36–41. doi: 10.1016/j.parepi.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro CJ, Morici G, Tomazic M, Vilte D, Bok M, Vega C, Parreño V, Schnittger L (2017) Prevalencia y patogenicidad de Cryptosporidium spp. en comparación con otros enteropatogenos de terneros de rodeos lecheros. II Jornadas de Investigación FCEQN UM, Moron, Argentina, 3 de Oct.
- Hamnes IS, Gjerde B, Robertson L. Prevalence of Giardia and Cryptosporidium in dairy calves in three areas of Norway. Vet Parasitol. 2006;140:204–216. doi: 10.1016/j.vetpar.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Henriksen SA, Pohlenz JFL. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet Scand. 1981;22:594–596. doi: 10.1186/BF03548684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Nacional de Estadística y Censos de la República Argentina (INDEC) (2008) Censo Nacional Agropecuario htpp: www.indec.gov.ar. (Accessed 09.07.16)
- Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013;141:1009–1020. doi: 10.1017/S0950268812001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo MM, Kirkland PD, Mohler VL, Perkins NR, Gunna AA, House JK. Prevalence of major enteric pathogens in Australian dairy calves with diarrhea. Aust Vet J. 2011;89(5):167–173. doi: 10.1111/j.1751-0813.2011.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupke A, Rzeżutka A. Emergence of novel subtypes of Cryptosporidium parvum in calves in Poland. Parasitol Res. 2015;114:4709–4716. doi: 10.1007/s00436-015-4719-1. [DOI] [PubMed] [Google Scholar]
- Kinross P, Beser J, Troell K, Silverlås C, Björkman C, Lebbad M, et al. Cryptosporidium parvum infections in a cohort of veterinary students in Sweden. Epidemiol Infect. 2015;143:2748–2756. doi: 10.1017/S0950268814003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque ASG, Zaidi AKM, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kváč M, Hromadová N, Kvetonová D, Rost M, Sak B. Molecular characterization of Cryptosporidium spp. in pre-weaned dairy calves in the Czech Republic: absence of C. ryanae and management-associated distribution of C. andersoni, C. bovis and C. parvum subtypes. Vet Parasitol. 2011;177:378–382. doi: 10.1016/j.vetpar.2010.11.048. [DOI] [PubMed] [Google Scholar]
- Mcguirk SM. Disease management of dairy calves and heifers. Vet Clin N Am Food Anim Pract. 2008;24:139–153. doi: 10.1016/j.cvfa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganck V, Hoflack G, Piepers S, Opsomer G. Evaluation of a protocol to reduce the incidence of neonatal calf diarrhoea on dairy herds. Prev Vet Med. 2015;118:64–70. doi: 10.1016/j.prevetmed.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic Z, Abe N. Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60kDa glycoprotein gene sequences. Parasitol. 2007;134:351–358. doi: 10.1017/S0031182006001508. [DOI] [PubMed] [Google Scholar]
- Olson ME, O’Handley RM, Ralston BJ, McAllister TA, Thompson RCA. Update on Cryptosporidium and Giardia infections in cattle. Trends Parasitol. 2004;20:185–191. doi: 10.1016/j.pt.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Peralta RH, Velásquez JN, Cunha FS, Pantano ML, Sodré FC, Silva S, Astudillo OG, Peralta JM, Carnevale S. Genetic diversity of Cryptosporidium identified in clinical samples from cities in Brazil and Argentina. Mem Inst Oswaldo Cruz. 2016;111:30–36. doi: 10.1590/0074-02760150303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutzer J, Karanis P. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet Parasitol. 2007;146:357–362. doi: 10.1016/j.vetpar.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Qi MZ, Fang YQ, Wang XT, Zhang LX, Wang RJ, Du SZ, et al. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, North-Western China. J Med Microb. 2015;64:111–116. doi: 10.1099/jmm.0.079327-0. [DOI] [PubMed] [Google Scholar]
- Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Sánchez C, Suero M, Castignani H, Terán JC, Marino M (2012) Estado actual y evolución de la lechería argentina (2008-2011). Reunión Anual de la Asociación Argentina de Economía Agraria. Corrientes. Argentina
- Santín M, Trout JM, Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol. 2008;155:15–23. doi: 10.1016/j.vetpar.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Silverlås C, Emanuelson U, de Verdier K, Bjorkman C. Prevalence and associated management factors of Cryptosporidium shedding in 50 Swedish dairy herds. Prev Vet Med. 2009;90:242–253. doi: 10.1016/j.prevetmed.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Silverlås C, Bosaeus-Reineck H, Näslund K, Björkman C. Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Inter J Parasit. 2013;43:155–161. doi: 10.1016/j.ijpara.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BB, Sharma R, Kumar H, Banga HS, Singh Aulakh R, Singh Gill JP, et al. Prevalence of Cryptosporidium parvum infection in Punjab (India) and its association with diarrhea in neonatal dairy calves. Vet Parasitol. 2006;140:162–165. doi: 10.1016/j.vetpar.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Smith RP, Clifton-Hadley FA, Cheney T, Giles M. Prevalence and molecular typing of Cryptosporidium in dairy cattle in England and Wales and examination of potential on-farm transmission routes. Vet Parasitol. 2014;204:111–119. doi: 10.1016/j.vetpar.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RCA, Ash A. Molecular epidemiology of Giardia and Cryptosporidium infections. Infec Gen Evolut. 2016;40:315–323. doi: 10.1016/j.meegid.2015.09.028. [DOI] [PubMed] [Google Scholar]
- Tiranti K, Larriestra A, Vissio C, Picco N, Alustiza F, Degioanni A, Vivas A. Prevalence of Cryptosporidium spp. and Giardia spp., spatial clustering and patterns of shedding in dairy calves from Córdoba, Argentina. R Bras Parasitol Vet. 2011;20:65–72. doi: 10.1590/S1984-29612011000200009. [DOI] [PubMed] [Google Scholar]
- Tomazic ML, Maidana J, Dominguez M, Louge Uriarte E, Galarza R, Garro C, et al. Molecular characterization of Cryptosporidium isolates from calves in Argentina. Vet Parasitol. 2013;198:382–386. doi: 10.1016/j.vetpar.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Trotz-Williams LA, Martin SW, Leslie KE, Duffield T, Nydam DV, Peregrine AS. Association between management practices and within-herd prevalence of Cryptosporidium parvum shedding on dairy farms in southern Ontario. Prev Vet Med. 2008;83:11–23. doi: 10.1016/j.prevetmed.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielinga PR, de Vries A, van der Goot TH, Mank T, Mars MH, Kortbeek LM, van der Giessen JWB. Molecular epidemiology of Cryptosporidium in humans and cattle in the Netherlands. Int J Parasitol. 2008;38:809–817. doi: 10.1016/j.ijpara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Xiao L, Fayer R. Molecular characterization of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunol Med Mic. 2008;52:309–323. doi: 10.1111/j.1574-695X.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- Xiao L, Escalante L, Yang C, Escalante AA, Montali RJ, Fayer R, Lal AA, Sulaiman I. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl EnvironMicrobiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KH, Bullock SL, Melvin DM, Spruill CL. Ethyl acetate as a substitute for diethyl ether in the formalin ether sedimentation technique. J Clin Microb. 1979;10:852–853. doi: 10.1128/jcm.10.6.852-853.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 17 kb)