Abstract
Cryptosporidium spp. in diarrheic calves less than 30 days old from farms across Northern Ireland were examined over a year period by microscopic, genotyping, and subtyping techniques to characterize the transmission dynamics. Cryptosporidium oocysts were detected in 291 of 779 (37.4%) animals. The prevalence rates of rotavirus, coronavirus, and Escherichia coli K99+ were lower as seen in 242 of 806 (30.0%), 46/806 (5.7%), and 16/421 (3.8%) of animals, respectively. Of the 224 Cryptosporidium-positive specimens available for molecular analysis, Cryptosporidium parvum was identified in 213 (95.1%) specimens, Cryptosporidium bovis in eight (3.6%), and Cryptosporidium deer-like genotype in three (1.3%). Sequence analysis of the 60-kDa glycoprotein gene identified 16 IIa subtypes and a new subtype family, with 120 of the 216 (55.6%) positive specimens having the subtype IIaA18G3R1. Eight of the IIa subtypes were previously seen in humans in Northern Ireland. Several subtypes were temporally or geographically unique. The genetic diversity in calves in Northern Ireland was much greater than that reported from other areas. This work demonstrates the utility of genotyping and subtyping tools in characterizing the transmission of Cryptosporidium spp. in calves and humans.
Keywords: Cryptosporidiosis, Cryptosporidium Infection, Santin, Neonatal Calf, Human Cryptosporidiosis
Introduction
Cattle production is the major agricultural sector in Northern Ireland (NI), producing over half of the annual agricultural output. The total cattle population in Northern Ireland in 2005 was 1.67 million animals, including 0.27 million animals less than 6 months of age. The total annual output in 2004 from all cattle operations was £752 million (DARD Statistical Data 1981–2005, http://www.dardni.gov.uk). Morbidity and mortality in cattle populations significantly reduce the income of cattle operations. One of the major causes of morbidity and mortality in cattle is diarrheal disease. One study reported that, of the total number of mortalities in calves less than 1 month of age in Northern Ireland in 1992, some 18.5% were due to gastrointestinal syndromes, a figure that rose to 48% when stillbirths were excluded (Menzies et al. 1996). Cryptosporidium, rotavirus, coronavirus, and Escherichia coli are the major enteric pathogens in neonatal calves of 30 days or younger (McDonough et al. 1994; Bendali et al. 1999; de la Fuente et al. 1999; Naciri et al. 1999).
Cattle are infected with at least four Cryptosporidium species, including Cryptosporidium parvum, Cryptosporidium bovis, Cryptosporidium andersoni, and the Cryptosporidium deer-like genotype (Xiao et al. 2004). A recent study in the United States has shown that the occurrence of these Cryptosporidium spp. in cattle is age-related (Santin et al. 2004). C. parvum is responsible for about 85% of the Cryptosporidium infections in preweaned calves but only 1% of the Cryptosporidium infections in postweaned calves. Postweaned calves and old cattle are mostly infected with C. bovis, C. andersoni, and the Cryptosporidium deer-like genotype (Santin et al. 2004). Nevertheless, C. parvum is the major species responsible for diarrhea in calves, and is the only zoonotic species in cattle.
Recently, researchers have used highly discriminatory subtyping techniques to study the transmission of bovine C. parvum infections in a geographic area (Mallon et al. 2003a,b; Tanriverdi et al. 2006; Tanriverdi and Widmer 2006). These tools are very useful in characterizing transmission dynamics of C. parvum in cattle, the maintenance of the parasite on cattle farms and the role of herd-to-herd transmission in epidemiology. One of the most commonly used subtyping tools is based on sequence analysis of the 60-kDa glycoprotein gene (GP60), which differentiates C. parvum to several subtype families and several subtypes within each family (Strong et al. 2000; Peng et al. 2001, 2003a,b; Sulaiman et al. 2001, 2005; Glaberman et al. 2002; Leav et al. 2002; Alves et al. 2003, 2006; Sturbaum et al. 2003; Wu et al. 2003; Zhou et al. 2003; Chalmers et al. 2005; Abe et al. 2006; Trotz-Williams et al. 2006). Of the two major C. parvum subtype families, the IIa subtype family is zoonotic in nature and is seen in both humans and calves, whereas the IIc subtype family is anthroponotic and is only found in humans (Alves et al. 2003; Xiao and Ryan 2004).
Currently, little is known about the transmission of Cryptosporidium spp. in cattle in Northern Ireland. A previous subtyping study of a small number of human specimens from outbreaks and sporadic cases identified several C. parvum IIa subtypes, suggesting that these zoonotic subtypes might be present in cattle (Glaberman et al. 2002). We report in this study a molecular epidemiologic study of cryptosporidiosis in neonatal calves in Northern Ireland.
Materials and methods
Sample collection
Fecal specimens submitted to the diagnostic laboratory at the Veterinary Sciences Division of the Department of Agriculture and Rural Development for Northern Ireland, Stormont, Belfast, were used in the study. They were collected from symptomatic calves less than 1 month of age on farms across Northern Ireland over a 12-month period during March 2002 to February 2003. They were examined for the presence of Cryptosporidium by a modified Ziehl–Neelsen staining technique (Baxby et al. 1984), rotavirus and coronavirus by enzymatic immunoassay using the Pathasure Enteritis Elisa Kit (Vetoquinol UK Ltd, Bicester, England), and enteropathogenic E. coli K99+ by bacterial culture on a defined minimal medium Minca agar (Guinee et al. 1976) followed by the use of the latex agglutination test FIMBREX K99 Kit (The Veterinary Laboratories Agency, Addlestone, Surrey, England).
DNA extraction and genotyping
Genomic DNA was extracted from Cryptosporidium-positive specimens by alkaline digestion and phenol–chloroform extraction, followed by DNA purification using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA), as previously described (Peng et al. 2003b). Cryptosporidium spp. presented were genotyped by nested polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis of an approximately 830-bp fragment of the small subunit (SSU) rRNA gene (Xiao et al. 2001). All secondary PCR products were sequenced to further confirm the genotype identification.
GP60 PCR
A fragment of the 60-kDa glycoprotein (GP60) gene of approximately 850 bp was amplified by nested PCR as previously described, using 1 or 2 μl of the extracted DNA in primary PCR (Alves et al. 2003). Secondary PCR products were purified using the Marligen Rapid PCR purification kit (Marligen Biosystems, Ijamsville, MD, USA). Sequencing reactions for the purified products were performed using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer’s instructions. SSU rRNA PCR products were sequenced in both directions using the forward and reverse primer used in secondary PCR, whereas GP60 products were mostly sequenced from the 5′ end. Unincorporated fluorescent-dye terminators from the cycle-sequencing reaction were removed using AutoSeq G-50 columns (Amersham Biosciences, Freiberg, Germany). Sequences were read on an ABI3100 automated sequencer (Applied Biosystems). The nucleotide sequences obtained in this study were aligned with reference sequences retrieved from the GenBank using the program ClustalX (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). The recently proposed nomenclature was used in naming C. parvum subtypes (Sulaiman et al. 2005).
Results
Prevalence of Cryptosporidium
Of the four common enteric pathogens in neonatal calves examined over 1 year period in this study, Cryptosporidium was the most prevalent, being detected in 291 of 779 (37.4%) animals by microscopy of fecal smears stained by the modified Ziehl–Neelsen staining technique. Rotavirus had a slightly lower prevalence rate, being found in 242 of 806 (30.0%) specimens tested. In contrast the prevalence rates of coronavirus (46/806 or 5.7% of animals) and E. coli K99+(16/421 or 3.8% of animals) were much lower (Table 1). A total of 224 specimens positive for Cryptosporidium by microscopy were available for molecular analysis. Among them, 62 specimens (27.6%) were concurrently positive for rotavirus, six (2.7%) for coronavirus, and three (1.3%) for E. coli K99+.
Table 1.
Prevalence of Cryptosporidium and other enteric pathogens in neonatal calves in Northern Ireland during March 2002 to February 2003
| Pathogen | Number of animals tested | Number of positives | Percent positive |
|---|---|---|---|
| Cryptosporidium spp. | 779 | 291 | 37.4 |
| Rotavirus | 806 | 242 | 30.0 |
| Coronavirus | 806 | 46 | 5.7 |
| E. coli K99+ | 421 | 16 | 3.8 |
Cryptosporidium infection was detected in calves in every month over the 1 year period. There were considerable monthly variations in infection rates. However, infection rates during the summer months were generally lower than those in other months (Fig. 1).
Fig. 1.
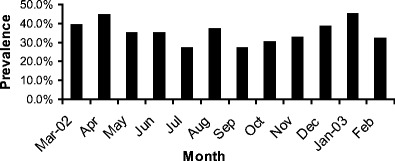
Monthly prevalence of Cryptosporidium spp. in calves less than 30 days old in Northern Ireland March 2002 to February 2003
Cryptosporidium genotypes
The 224 specimens positive for Cryptosporidium by microscopy and available for molecular analysis were collected from neonatal calves on 193 farms across Northern Ireland. DNA extracted from them all yielded SSU rRNA gene products of the expected size in nested PCR. RFLP analysis with SspI and VspI produced similar SspI banding patterns and identical VspI banding pattern for all PCR products. The SspI RFLP pattern of most PCR products was indicative of C. parvum, which were confirmed by results of DNA sequencing. However, a few SspI RFLP products yielded an upper band and a lower band that were slightly smaller than PCR products of C. parvum. DNA sequencing indicated that they were C. bovis or Cryptosporidium deer-like genotype. A few PCR products had a combination of the regular and the smaller upper bands. DNA sequencing indicated that they were C. bovis (Fig. 2). Altogether, C. parvum was identified in 213 specimens, C. bovis in eight specimens and Cryptosporidium deer-like genotype in three specimens. The SSU rRNA sequences of C. parvum, C. bovis and the deer-like genotype were identical to those from these species previously reported in the United States (Xiao et al. 2002; Santin et al. 2004).
Fig. 2.
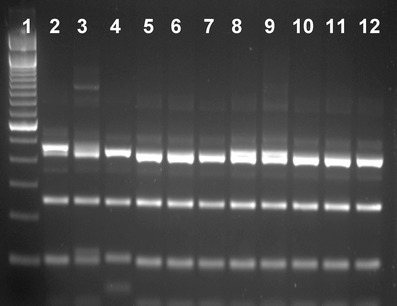
SspI RFLP patterns of three Cryptosporidium species in neonatal calves. Lane 1 100-bp molecular markers; lanes 2, 3, 5–9: C. bovis; lane 4: C. parvum; lanes 10–12: Cryptosporidium deer-like genotype. Note the upper and lower bands of C. parvum (lane 4) are slightly larger than those of the deer-like genotype (lanes 10–12) and some C. bovis (lanes 5–7), and some C. bovis (lanes 2, 3, 8, and 9) have a mixture of the larger and smaller upper bands
GP60 subtypes
Of the 224 specimens analyzed by GP60 PCR, 216 produced products of the expected size, including all 213 specimens identified as C. parvum by SSU rRNA PCR and three of the eight specimens identified as C. bovis. These products were sequenced successfully, except for five, which had sequence ambiguity at the 5′ end. Alignment of the sequences obtained with reference sequences revealed that 215 sequences belonged to the C. parvum subtype family IIa, whereas the remaining sequence obtained from one C. parvum-infected calf had less than 90% homology to other sequences, thus belonged to a new C. parvum subtype family. Altogether, 16 IIa subtypes were found in neonatal calves studied (Fig. 3), which differed from each other only in the number of TCA (designated by the letter A) and TCG (designated by the letter G) repeats. All IIa subtype sequences had only one copy of the ACATCA sequence (designated by the letter R). The most common subtype was IIaA18G3R1, which was found in 120 calves. Other less common subtypes included IIaA15G2R1, IIaA17G2R1, IIaA19G4R1, IIaA20G3R1, IIaA19G3R1, IIaA17G3R1, IIaA20G5R1, IIaA18G2R1, and IIaA20G2R1, which were seen in 28, 19, 15, 6, 5, 5, 3, 2, and 2 calves, respectively. The new subtype family and other IIa subtypes (IIaA16G3R1, IIaA17G1R1, IIaA18R1, IIaA19G2R1, IIaA20G4R1, and IIaA21G2R1) were each found in only one animal. All five sequences with 5′ end ambiguity belonged to IIa subtype family (Fig. 3).
Fig. 3.
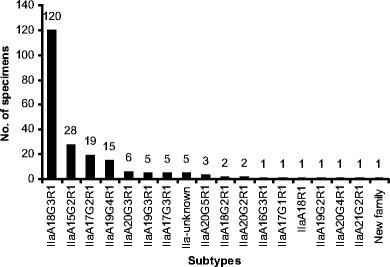
Distribution of Cryptosporidium parvum subtypes in neonatal calves in Northern Ireland
Distribution of subtypes
Among the four more common IIa subtypes, IIa18G3R1 was found year around in all geographic areas in Northern Ireland (Fig. 4). Subtype IIaA15G2R1, though much less common, was also found at low frequency in all areas, with the exception of a high occurrence in April 2002. Subtype IIaA17G2R1 also mainly occurred in April 2002, but this was mostly due to the more extensive sampling of animals on a farm in the Lough Neagh basin area during an outbreak of cryptosporidiosis, in which all positive animals were infected with the subtype. Subtype IIaA19G4R1 was found sporadically over the year (Fig. 4), predominantly in animals within a 15-mile radius in the North Antrim area, suggesting the possibility of some common point of infection or transmission.
Fig. 4.
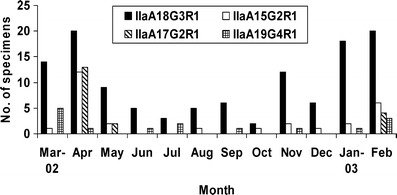
Monthly distribution of four common Cryptosporidium parvum subtypes in neonatal calves in Northern Ireland
Discussion
Previous studies in other countries have shown Cryptosporidium, rotavirus, coronavirus, and enterotoxigenic E. coli are some common causes of diarrhea in neonatal calves less than 30 days old (Reynolds et al. 1986; Holland 1990; McDonough et al. 1994; de la Fuente et al. 1998; Bendali et al. 1999; Naciri et al. 1999). Results of this study suggest that the same is also true in Northern Ireland. The high prevalence of Cryptosporidium and rotavirus in neonatal calves in the current study is in agreement with similar studies conducted in Spain and France, which also showed lower prevalence of coronavirus and enterotoxigenic E. coli than the other two pathogens (de la Fuente et al. 1998, 1999; Naciri et al. 1999; Garcia et al. 2000). Similar to what was observed before (Moore and Zeman 1991; Garcia et al. 2000), concurrent Cryptosporidium and rotavirus infections were common in the present study.
The higher occurrence of Cryptosporidium infections in neonatal calves during winter and spring months is consistent with farming practice in Northern Ireland. Because of the adverse climatic conditions prevailing in Northern Ireland during the winter months of October through to March, most stock is housed indoors during this period. This is accompanied by a concomitant increase in the numbers of susceptible calves being born while cows are housed indoors. This potentially provides ideal conditions for the transmission of Cryptosporidium in newborn calves, either from direct contact with infected animals, or from contaminated bedding or housing, an infection source previously identified (Atwill et al. 1998). The lower detection rates during summer months were probably due to reductions in the numbers of symptomatic animals as a result of turnout of calves to pasture and a seasonal reduction in the numbers of calves being born. Similar patterns of spring and winter peaks have previously been seen in the United States (Xiao and Herd 1994; Mohammed et al. 1999).
The genotype distribution of Cryptosporidium seen in neonatal calves is very similar to that recently reported in the United States, with most infections due to C. parvum and C. bovis and the deer-like genotype only occasionally seen in neonatal calves younger than 30 days (Santin et al. 2004). The conservative nature of the SSU rRNA gene sequences for the three species in different geographic areas indicates that the age-related infections with different Cryptosporidium spp. previously observed in the United State probably also occurs in cattle in Northern Ireland.
The existence of 17 C. parvum subtypes may be indicative of a high genetic diversity of C. parvum in Northern Ireland. Previous studies in Portugal, United States and Canada have identified only a few C. parvum GP60 subtypes in calves in each area (Alves et al. 2003, 2006; Peng et al. 2003b; Trotz-Williams et al. 2006). It is not clear whether the higher parasite diversity seen in this study was due to more extensive sampling of many farms. Previously, a multilocus typing study also demonstrated the existence of many subtypes in calves in Scotland (Mallon et al. 2003b). The existence of several subtypes in a small area in theory can be the result of infected animals being brought into the area, either from local, national or international sources, and introduction of these new subtypes to the native cattle population. However, the majority of calves on Northern Ireland farms are either home-bred or bought in from local sources such as livestock marts, so it is more likely the high genetic diversity of the parasite seen here represents geographic isolation due to reduced movement of animals between farms.
Over half of the animals examined in Northern Ireland were infected with the subtype IIaA18G3R1. This subtype was previously seen in two waterborne outbreaks of human cryptosporidiosis in Northern Ireland in 2000 and 2001 (Glaberman et al. 2002). It was also seen in two sporadic human cases in Australia and one calf in Canada (Chalmers et al. 2005; Trotz-Williams et al. 2006). The widespread distribution both geographically and temporally indicates the successful transmission capabilities of this subtype either by direct animal-to-animal contact or through environmental contamination. It is interesting to note that another more widely distributed C. parvum subtype, IIaA15G2R1, was only seen in 13% of infected calves. The latter was responsible for 85% of C. parvum infections in calves and other ruminants in Portugal (Alves et al. 2006), and has been found in humans and calves in the United States, Canada, United Kingdom, Portugal, Slovenia, Australia, Japan, and Kuwait (Glaberman et al. 2002; Peng et al. 2003b; Stantic-Pavlinic et al. 2003; Wu et al. 2003; Chalmers et al. 2005; Sulaiman et al. 2005; Abe et al. 2006; Alves et al. 2006; Trotz-Williams et al. 2006).
In addition to IIaA18G3R1 and IIaA15G2R1, six other subtypes found in calves in the present study, including IIaA17G2R1, IIaA20G3R1, IIaA19G3R1, IIaA17G3R1, IIaA19G2R1, and IIaA20G4R1, were previously reported in humans in Northern Ireland (Glaberman et al. 2002). Another subtype in calves, IIaA17G1R1, has been found in three outbreaks of cryptosporidiosis in neighboring England and Wales that were associated with direct contact with animals or consumption of contaminated water (Chalmers et al. 2005). The similar distribution of C. parvum subtypes between humans and calves in the same geographic area suggests a role for calves in the transmission of human cryptosporidiosis in this area, even though more studies are clearly needed to assess the extent of calf-to-human transmission. As expected, the anthroponotic C. parvum IIc subtype family, which is the most common type of C. parvum in humans in most countries (Alves et al. 2003; Xiao et al. 2003; Xiao and Ryan 2004), was not seen in calves in the current and in previous studies.
In conclusion, results of the study have shown the occurrence of C. bovis and the Cryptosporidium deer-like genotype in the calves in Northern Ireland and the uniqueness of C. parvum transmission on farms in Northern Ireland, with one predominant subtype in neonatal calves. The widespread occurrence of the subtype in calves and humans demonstrates the genetic fitness of the parasite in the transmission of cryptosporidiosis. It also raises some concerns about the selection of highly infectious subtypes by intensive animal husbandry. Future multilocus subtyping studies would increase our understanding of the transmission dynamics of C. parvum in cattle and humans.
Acknowledgements
H.T. was funded by a Research Studentship from the Department of Agriculture and Rural Development for Northern Ireland. C.J.L. was supported by an E.U. Framework V (Quality of Life) Grant (PLK1-CT-1999-00775). The authors thank Prof. W.H.I. McLean, University of Dundee, Scotland, for provision of sequencing facilities. Ms. Velma Hayes, University of Ulster and the staff of the Diagnostic Unit of the Stormont Laboratory of the Veterinary Sciences Laboratory of the Department of Agriculture and Rural Development of Northern Ireland for technical assistance.
Footnotes
Nucleotide sequence data reported in this paper are available in the GenBank database under the accession numbers DQ648531-DQ648547. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- Abe N, Matsubayashi M, Kimata I, Iseki M. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res. 2006;99:303–305. doi: 10.1007/s00436-006-0140-0. [DOI] [PubMed] [Google Scholar]
- Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M, Xiao L, Antunes F, Matos O. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol Res. 2006;99:287–292. doi: 10.1007/s00436-006-0164-5. [DOI] [PubMed] [Google Scholar]
- Atwill ER, Harp JA, Jones T, Jardon PW, Checel S, Zylstra M. Evaluation of periparturient dairy cows and contact surfaces as a reservoir of Cryptosporidium parvum for calfhood infection. Am J Vet Res. 1998;59:1116–1121. [PubMed] [Google Scholar]
- Baxby D, Blundell N, Hart CA. The development and performance of a simple, sensitive method for the detection of Cryptosporidium oocysts in faeces. J Hyg (Lond) 1984;93:317–323. doi: 10.1017/s0022172400064858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendali F, Bichet H, Schelcher F, Sanaa M. Pattern of diarrhoea in newborn beef calves in south-west France. Vet Res. 1999;30:61–74. [PubMed] [Google Scholar]
- Chalmers RM, Ferguson C, Caccio S, Gasser RB, Abs El-Osta YG, Heijnen L, Xiao L, Elwin K, Hadfield S, Sinclair M, Stevens M. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int J Parasitol. 2005;35:397–410. doi: 10.1016/j.ijpara.2005.01.001. [DOI] [PubMed] [Google Scholar]
- de la Fuente R, Garcia A, Ruiz-Santa-Quiteria JA, Luzon M, Cid D, Garcia S, Orden JA, Gomez-Bautista M. Proportional morbidity rates of enteropathogens among diarrheic dairy calves in central Spain. Prev Vet Med. 1998;36:145–152. doi: 10.1016/S0167-5877(98)00077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente R, Luzon M, Ruiz-Santa-Quiteria JA, Garcia A, Cid D, Orden JA, Garcia S, Sanz R, Gomez-Bautista M. Cryptosporidium and concurrent infections with other major enterophatogens in 1 to 30-day-old diarrheic dairy calves in central Spain. Vet Parasitol. 1999;80:179–185. doi: 10.1016/S0304-4017(98)00218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A, Ruiz-Santa-Quiteria JA, Orden JA, Cid D, Sanz R, Gomez-Bautista M, de la Fuente R. Rotavirus and concurrent infections with other enteropathogens in neonatal diarrheic dairy calves in Spain. Comp Immunol Microbiol Infect Dis. 2000;23:175–183. doi: 10.1016/S0147-9571(99)00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaberman S, Moore JE, Lowery CJ, Chalmers RM, Sulaiman I, Elwin K, Rooney PJ, Millar BC, Dooley JS, Lal AA, Xiao L. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg Infect Dis. 2002;8:631–633. doi: 10.3201/eid0806.010368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinee PA, Jansen WH, Agterberg CM. Detection of the K99 antigen by means of agglutination and immunoelectrophoresis in Escherichia coli isolates from calves and its correlation with entertoxigenicity. Infect Immun. 1976;13:1369–1377. doi: 10.1128/iai.13.5.1369-1377.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland RE. Some infectious causes of diarrhea in young farm animals. Clin Microbiol Rev. 1990;3:345–375. doi: 10.1128/cmr.3.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leav BA, et al. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect Immun. 2002;70:3881–3890. doi: 10.1128/IAI.70.7.3881-3890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon M, MacLeod A, Wastling J, Smith H, Reilly B, Tait A. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J Mol Evol. 2003;56:407–417. doi: 10.1007/s00239-002-2412-3. [DOI] [PubMed] [Google Scholar]
- Mallon ME, MacLeod A, Wastling JM, Smith H, Tait A. Multilocus genotyping of Cryptosporidium parvum Type 2: population genetics and sub-structuring. Infect Genet Evol. 2003;3:207–218. doi: 10.1016/S1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- McDonough SP, Stull CL, Osburn BI. Enteric pathogens in intensively reared veal calves. Am J Vet Res. 1994;55:1516–1520. [PubMed] [Google Scholar]
- Menzies FD, Bryson DG, McCallion T, Matthews DI. Mortality in cattle up to two years old in Northern Ireland during 1992. Vet Rec. 1996;138:618–622. doi: 10.1136/vr.138.25.618. [DOI] [PubMed] [Google Scholar]
- Mohammed HO, Wade SE, Schaaf S. Risk factors associated with Cryptosporidium parvum infection in dairy cattle in southeastern New York State. Vet Parasitol. 1999;83:1–13. doi: 10.1016/S0304-4017(99)00032-1. [DOI] [PubMed] [Google Scholar]
- Moore DA, Zeman DH. Cryptosporidiosis in neonatal calves: 277 cases (1986–1987) J Am Vet Med Assoc. 1991;198:1969–1971. [PubMed] [Google Scholar]
- Naciri M, Lefay MP, Mancassola R, Poirier P, Chermette R. Role of Cryptosporidium parvum as a pathogen in neonatal diarrhoea complex in suckling and dairy calves in France. Vet Parasitol. 1999;85:245–257. doi: 10.1016/S0304-4017(99)00111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, Sulaiman IM, Glaberman S, Lal AA, Xiao L (2001) A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol :28S–31S [DOI] [PubMed]
- Peng MM, Meshnick SR, Cunliffe NA, Thindwa BD, Hart CA, Broadhead RL, Xiao L. Molecular epidemiology of cryptosporidiosis in children in Malawi. J Eukaryot Microbiol. 2003;50(Suppl):557–559. doi: 10.1111/j.1550-7408.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Peng MM, Wilson ML, Holland RE, Meshnick SR, Lal AA, Xiao L. Genetic diversity of Cryptosporidium spp. in cattle in Michigan: implications for understanding the transmission dynamics. Parasitol Res. 2003;90:175–180. doi: 10.1007/s00436-003-0843-4. [DOI] [PubMed] [Google Scholar]
- Reynolds DJ, Morgan JH, Chanter N, Jones PW, Bridger JC, Debney TG, Bunch KJ. Microbiology of calf diarrhoea in southern Britain. Vet Rec. 1986;119:34–39. doi: 10.1136/vr.119.2.34. [DOI] [PubMed] [Google Scholar]
- Santin M, Trout JM, Xiao L, Zhou L, Greiner E, Fayer R. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol. 2004;122:103–117. doi: 10.1016/j.vetpar.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Stantic-Pavlinic M, Xiao L, Glaberman S, Lal AA, Orazen T, Rataj-Verglez A, Logar J, Berce I. Cryptosporidiosis associated with animal contacts. Wien Klin Wochenschr. 2003;115:125–127. doi: 10.1007/BF03040292. [DOI] [PubMed] [Google Scholar]
- Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15-and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–4134. doi: 10.1128/IAI.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturbaum GD, Jost BH, Sterling CR. Nucleotide changes within three Cryptosporidium parvum surface protein encoding genes differentiate genotype I from genotype II isolates. Mol Biochem Parasitol. 2003;128:87–90. doi: 10.1016/S0166-6851(03)00017-3. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Lal AA, Xiao L (2001) A population genetic study of the Cryptosporidium parvum human genotype parasites. J Eukaryot Microbiol :24S–27S [DOI] [PubMed]
- Sulaiman IM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriverdi S, Widmer G. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect Genet Evol. 2006;6:113–122. doi: 10.1016/j.meegid.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S, Markovics A, Arslan MO, Itik A, Shkap V, Widmer G. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl Environ Microbiol. 2006;72:2507–2513. doi: 10.1128/AEM.72.4.2507-2513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotz-Williams LA. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res. 2006;99:346–352. doi: 10.1007/s00436-006-0157-4. [DOI] [PubMed] [Google Scholar]
- Wu Z, Nagano I, Boonmars T, Nakada T, Takahashi Y. Intraspecies polymorphism of Cryptosporidium parvum revealed by PCR-restriction fragment length polymorphism (RFLP) and RFLP-single-strand conformational polymorphism analyses. Appl Environ Microbiol. 2003;69:4720–4726. doi: 10.1128/AEM.69.8.4720-4726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Herd RP. Infection pattern of Cryptosporidium and Giardia in calves. Vet Parasitol. 1994;55:257–262. doi: 10.1016/0304-4017(93)00645-F. [DOI] [PubMed] [Google Scholar]
- Xiao L, Ryan UM. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis. 2004;17:483–490. doi: 10.1097/00001432-200410000-00014. [DOI] [PubMed] [Google Scholar]
- Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH, Lal AA. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- Xiao L, Sulaiman IM, Ryan UM, Zhou L, Atwill ER, Tischler ML, Zhang X, Fayer R, Lal AA. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int J Parasitol. 2002;32:1773–1785. doi: 10.1016/S0020-7519(02)00197-2. [DOI] [PubMed] [Google Scholar]
- Xiao L, Bern C, Sulaiman IM, Lal AA. Molecular epidemiology of human cryptosporidiosis. In: Thompson RCA, Armson A, Ryan UM, editors. Cryptosporidium: from molecules to disease. Amsterdam: Elsevier; 2003. pp. 121–146. [Google Scholar]
- Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Singh A, Jiang J, Xiao L. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol. 2003;41:5254–5257. doi: 10.1128/JCM.41.11.5254-5257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


