Abstract
The diversity and function of viruses in coral holobionts has only recently received attention. The non-reef building gorgonian octocoral, Gorgonia ventalina, is a major constituent of Caribbean reefs. We investigated viral communities associated with G. ventalina tissues to understand their role in gorgonian ecology. Pyrosequencing was used to prepare a total of 514,632 sequence reads of DNA- and RNA-based mixed-community viral genomes (metaviromes). RNA viral assemblages were comprised of primarily unidentifiable reads, with most matching host transcripts and other RNA metaviromes. DNA metaviromes were similar between healthy and diseased tissues and comprised of contiguous sequences (contigs) that matched primarily metazoan and bacterial proteins. Only ~5% of contigs matched viral proteins that were primarily cyanophage and viruses of Chlorella and Ostreococcus. Our results confirm that DNA and RNA viruses comprise a component of the gorgonian holobiont, suggesting that they may play a role in the ecology of G. ventalina.
Keywords: Virus, Metavirome, Gorgonian, Community
Introduction
Sea fans (Anthozoa, Octocorallia, and Gorgonaceae) are non-reef building corals found throughout the world, but with highest densities in the Caribbean Sea. Although sea fans are major constituents of Caribbean reefs, there have been few cultivation-independent studies characterizing the composition of gorgonian-associated microorganisms (Sunagawa et al. 2010). Furthermore, studies of microbe-sea fan interactions have focused primarily on the fungal disease aspergillosis, putatively caused by the pathogen, Aspergillus sydowii (Smith et al. 1996). The viral component of the coral holobiont has only recently been studied. Virus-like particles have been observed using TEM and microscopic-based approaches (Wilson et al. 2005; Davy and Patten 2007), and UV-inducible virus-like particles were observed in zooxanthellae symbiotic with anemones (Wilson et al. 2001) and corals (Lohr et al. 2007). The advent of high throughput sequencing has permitted comprehensive studies of viral assemblages in seawater (Breitbart et al. 2002) and sediments (Breitbart et al. 2004), and in association with corals and other metazoans (Thurber et al. 2008; Ng et al. 2009). Metagenomic studies of coral-associated viruses have focused exclusively on scleractinian corals (Wegley et al. 2007; Marhaver et al. 2008; Thurber et al. 2008).
There have been no previous investigations of viruses in gorgonian corals. The aim of this survey was to establish the presence and identity of viral assemblages associated with the sea fan Gorgonia ventalina, which dominates reef sea fan communities in the Caribbean. We used a metagenomic approach to elucidate the composition of RNA and DNA viruses associated with tissues. Our data suggest that viral assemblages associated with gorgonians contain a wide suite of viral genotypes, and that healthy and diseased tissues harbor similar viral complements.
Methods
Sampling location and collection
Samples for metaviromic analysis were collected by SCUBA at Wreck Reef, La Parguera, Puerto Rico (17°56.093 N, 67°02.931 W) in May 2009. Two × 2 cm fragments of Gorgonia ventalina were collected from each of six apparently healthy and five putatively aspergillosis-affected colonies. Fragments were placed in Whirlpaks©, then immediately transported to shore, flash frozen in liquid nitrogen, stored at −80°C, and shipped to Cornell University on dry ice.
Metaviromic library preparation
Ten samples of sea fans (5 each of visually ‘healthy’ and ‘diseased’ tissues) were used to generate metagenomic libraries of the viral fraction following protocols for tissues in Thurber et al. (2009). The sea fan samples were first pooled then homogenized in 0.02 μm filtered PBS (pH 7.4). The homogenate was then centrifuged to pellet cells, filtered, treated with chloroform, and subjected to CsCl density gradient centrifugation as described by Thurber and colleagues. The fraction between the 1.3 and 1.5 g ml−1 was used in order to target most dsDNA and some RNA bacteriophage and viruses (Thurber et al. 2009). Viral DNA was treated with DNAse I prior to viral nucleic acid extraction and purification as described. Attempted PCR amplification of 16S and 18S rRNAs from purified viruses (Thurber et al. 2009) did not yield amplicons. Viral RNA was isolated by first treating a split of concentrated and purified viruses with 10 μl RNAse A (2 mg ml−1), which was incubated for 2 h at 37°C to eliminate free RNAs. The RNA as then extracted using the RNEasy Plus Kit (Qiagen) following the Thurber et al. (2009) protocol. DNA viruses were amplified prior to sequences by multiple displacement amplification (MDA) using the Genomiphi© (GE Healthcare) kit. Triplicate amplifications were performed on 1 μl viral DNA, which were then combined, and purified using the DNEasy Tissue kit (Qiagen). RNA viruses were amplified using the Transplex Whole Transcriptome Amplification 2 kit (Sigma) following manufacturer’s recommendations, where reactions were cleaned using the DNA Clean and Concentrator-5 kit (Zymo Research). DNA was sequenced at the Broad Institute (MIT) using Titanium 454 Pyrosequencing (1/8th of a plate per library). RNA converted to ds cDNA was subjected to 454 Titanium Pyrosequencing at the EnGenCore laboratory (University of South Carolina; 1/8th of a plate per library). Sequence information generated in this study has been deposited at CAMERA (http://camera.calit2.net) under project BroadPhageMetagenomes for DNA viruses and CAM_P_0000792 for RNA Viruses.
Metagenome analysis
Sequence libraries were initially imported into the CLC Genomics Workbench 4.0, where they were trimmed of MID tags, low-quality sequences, and then subjected to de novo assembly (minimum overlap 20% of length, minimum nucleotide identity 0.95) into contiguous fragments (contigs). Because of the potential for the metaviromes to contain putative host nucleic acids that were inadvertently collected as part of the metavirome preparation, metaviromic libraries were compared by BLASTn to a transcriptome sequenced from G. ventalina tissues where matching sequences at e-values < 1 e−10 were eliminated from further analyses. Non-host sequence reads within RNA virus libraries and contiguous sequences >100 bp in the DNA virus libraries were compared against the non-redundant (nr) protein database by BLASTx. Contiguous sequences > 5 kBp were selected for further analysis in all libraries because of greatest confidence in annotation. Open reading frames (ORFs) on all large contigs > 5 kBp in each library were identified using the GETORF algorithm. Large contig ORFs were first compared to the nr database at NCBI using the BLASTx algorithm to identify putatively bacteriophage or viral contigs (i.e., those which had >1 ORF matching at e-values < 10−5 to viral proteins). Putatively viral contigs were then subject to reciprocal BLASTn analyses between libraries to establish contigs unique to the diseased DNA virus library. Contigs > 100 bp that did not match protein databases by BLASTx against nr at e-values < 10−5 were further characterized by tBLASTx analysis against all viral genomes in GenBank. Individual RNA virus reads not matching proteins in the non-redundant database were further compared against an RNA metavirome prepared from Lake Needwood, MD (Djikeng et al. 2009) by tBLASTx analysis.
Results and discussion
Metaviromic libraries were comprised of a total of 514,632 sequences (Table 1). The proportion of annotated sequence reads (BLASTx against nr databse) and contigs was highest for DNA virus libraries and least for the RNA viral reads (Fig. 1). Pairwise reciprocal BLASTn analysis between libraries revealed substantial overlap in total sequence space between HV and DV DNA libraries (15.4% of all sequence reads were matched at e < 0.001) and RNA libraries (35.4%), but only minimal overlap between RNA and DNA libraries (0.1–0.5% of total sequences). The majority of matches for both DNA and RNA viral contigs and reads were to bacterial and metazoan proteins. The remaining sequences mostly did not match sequenced proteins of any group (~89%). RNA metagenomic hits to viral proteins (0.1–0.2% of sequences) shared only weak homology to bacteriophage to Chlorella virus (a ds DNA virus in the Phycodnaviridae), and to Bat SARS Coronavirus (ssRNA). Comparison to the Silva database (Pruesse et al. 2007) revealed only 10 HV and 9 DV RNA contigs sharing nucleotide identity to rRNAs. We have several reasons to believe that the unknown viral reads in RNA libraries represent naturally occurring viruses. Firstly, a total of 549 HV and 562 DV RNA library reads matched at E < 0.001 by tBLASTx to a freshwater lake RNA metavirome (Djikeng et al. 2009). Of these, 442 HV and 429 DV RNA library reads shared no homology with sequenced proteins. Secondly, the low G + C percentage of both HV and DV libraries is typical of RNA viruses (Culley et al. 2007) and is substantially lower than environmental mRNAs recovered in metatranscriptomic surveys targeting bacteria (Hewson et al. 2010). Our low percentage of annotateable RNA viral sequences is in line with previous viral metaviromic surveys (Breitbart et al. 2002, 2003, 2004; Angly et al. 2006; Culley et al. 2006; Bench et al. 2007; Schoenfeld et al. 2008; Srinivasiah et al. 2008; Djikeng et al. 2009) and reflects the poor state of knowledge of RNA viruses in marine ecosystems.
Table 1.
Library characteristics of healthy viral (HV) and diseased viral (DV) DNA and RNA metagenomic libraries prepared from G. ventalina
| Parameter | DNA | RNA | ||
|---|---|---|---|---|
| HV | DV | HV | DV | |
| Reads in library | 222,787 | 258,137 | 16,630 | 17,078 |
| Hits to host transcriptome | 10,156 | 2,091 | 8,571 | 8,955 |
| Reads assembling to contigs | 193,501 | 221,940 | 12,544 | 12,943 |
| Contigs > 100 bp | 19,885 | 13,308 | 133 | 117 |
| Contigs > 500 bp | 13,442 | 7,849 | 17 | 6 |
| Contigs > 5 kBp | 134 | 30 | 0 | 0 |
| Average Length (bp) | 381 | 358 | 143 | 143 |
| Average G + C % | 52.6 | 45.4 | 37.3 | 37.8 |
| Non-host matches to nr protein database | 11,264 | 5,874 | 842 | 1,084 |
nr = Non-redundant protein database at NCBI. Contigs = contiguous sequences generated using CLC Genomics Workbench 4.0 (minimum overlap = 0.2 and percent ID = 95%)
Fig. 1.
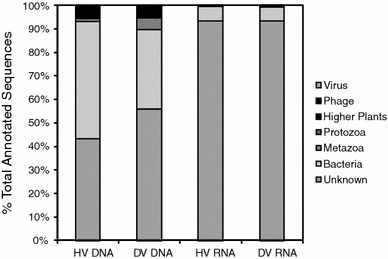
Annotation of HV and DV DNA contiguous contigs > 100 bp and HV and DV RNA reads. Best matches to the non-redundant (nr) protein database at NCBI were examined using the BLASTx algorithm (Altschul et al. 1997) with an e-value cutoff of 10−3. The phylogeny of annotated proteins was examined by Uniprot Taxonomy. Total sequences are indicated in Table 1
In contrast to the RNA metaviromes, DNA viruses were well annotated, where ~50% of contigs > 100 bp matched proteins in the nr database. However, sequences matching viral proteins comprised only a small percentage of both HV and DV libraries (5.1 and 4.8%, respectively), and both libraries were dominated by sequences that matched bacterial proteins. The best viral or bacteriophage matches for DNA libraries were to cyanophage of Synechococcus and Prochlorococcus. The large number of putatively bacterial sequences and dominance of cyanophage sequences may reflect biases in the total number of sequenced phage and bacterial genomes in genome databases.
HV DNA contigs > 5 kBp yielded 11,550 ORFs, while DV large contigs yielded 2,596 ORFs. BLASTx comparison of HV and DV ORFs against the non-redundant protein database at NCBI elucidated 18 HV and 17 DV large contigs, which had ≥ 1 ORF that matched viral proteins. Of these, 7 contigs from each of the DV and HV contigs had multiple ORFs annotated as viral or phage proteins. Of those large contigs with multiple strong ORF matches (E-value < 10−10) to viral proteins, several matched cyanophage (2 each in HV and DV libraries), with the remainder matching bacteriophage of heterotrophic bacteria (Fig. 2). The gorgonian holobiont is comprised of eukaryotic microorganisms (notably symbiotic dinoflagellates and fungi) and bacteria (Ribes et al. 1998; Toledo-Hernandez et al. 2007); cyanobacteria have not yet been reported from their tissues. However, given they occur as members of the holobiont in other corals (Lesser et al. 2004; Wegley et al. 2007), it is likely that they occur in gorgonians. In both HV and DV DNA viruses, most viral ORF matches were to conserved hypothetical proteins or proteins of unknown function (~52% for both libraries). Remaining viral ORFs were comprised of structural proteins (20%) and replication-associated proteins (9%) for the HV contigs, and structural proteins (14%), integrases (12%), and helicases (10%) among DV contigs. There were no recognizable metazoan viruses among DNA metaviromes. This may be because metazoan viruses that occur within the gorgonian holobiont are not well represented in sequence databases, that they share only weak homology to sequenced metazoan viral genomes, or they were not present in the buoyancy fraction used to prepare metaviromes (1.3–1.5 g ml−1).
Fig. 2.
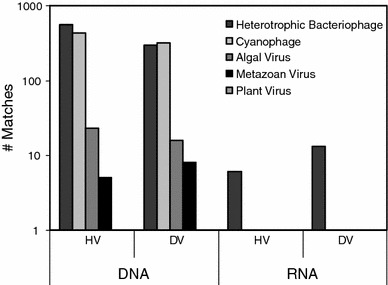
Phage and viral matches of HV and DV DNA contigs and RNA reads to the non-redundant protein database at NCBI. The phylogeny of annotated proteins was examined by Uniprot taxonomy and is based on BLASTx comparison with an e-value cutoff of 10−3
The gorgonian holobiont contains a microbial community that is known to be comprised of the cnidarian host, zooxanthellae, bacteria, fungi, and slime molds (Smith et al. 1996; Gil-Agudelo et al. 2006; Toledo-Hernandez et al. 2007; Sunagawa et al. 2010). Our study extends the concept of the gorgonian holobiont to viruses. We successfully surveyed RNA and DNA viruses from both healthy and diseased sea fan tissues. These viruses may play a similar role within the holobiont as in seawater, causing microbial mortality (Proctor and Fuhrman 1990), shunting organic matter into labile forms that are taken up rapidly by uninfected bacteria (Middelboe et al. 1996), and regulating the abundance of dominant groups of bacteria (Thingstad and Lignell 1997). This work involved significant new viral discovery because most nucleotide sequence information generated in this study does not match known proteins or genomes. Despite our inability to affiliate most metagenomic information with viral taxa, we detected substantial overlap in sequence space between healthy and diseased DNA and RNA viral assemblages (but not between RNA to DNA viruses). Nucleic acid sequences matching known viruses provide insight into putative hosts that may form part of the gorgonian holobiont, which include groups that are well represented in plankton.
Acknowledgments
The authors are grateful to the Gordon and Betty Moore Foundation Viral Sequencing Initiative for DNA metaviromic sequencing. The authors thank N. Douglas, D. Doud, S. Gitlin and J. Barbosa for assistance in conducting laboratory preparation of metaviromes and E. Weil for assistance in sample collection. Helpful comments on interpretation of metaviromes were provided by E. Wommack. This work was supported by NSF grants OCE-0961894, OCE-1049670, DEB-1028898 and Federal Formula Funds grant NYS-189489 awarded to IH, OCE-0849799 to CDH, and the Cornell Biogeochemistry and Environmental Biocomplexity Initiative.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy J, Mueller J, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:2121–2131. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, Wommack KE. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Felts B, Kelley S, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Diversity and population structure of a near-shore marine-sediment viral community. Proc R Soc Lond B. 2004;271:565–574. doi: 10.1098/rspb.2003.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J. 2007;4:69. doi: 10.1186/1743-422X-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy JE, Patten NL. Morphological diversity of virus-like particles within the surface microlayer of scleractinian corals. Aquat Microb Ecol. 2007;47:37–44. doi: 10.3354/ame047037. [DOI] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One. 2009;4:e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Agudelo DL, Myers C, Smith GW, Kim K. Changes in the microbial communities associated with Gorgonia ventalina during aspergillosis infection. Dis Aquat Org. 2006;69:89–94. doi: 10.3354/dao069089. [DOI] [PubMed] [Google Scholar]
- Hewson I, Poretsky RS, Tripp HJ, Montoya JP, Zehr JP. Spatial patterns and light-driven variation of microbial population gene expression in surface waters of the oligotrophic open ocean. Environ Microbiol. 2010;12:1940–1956. doi: 10.1111/j.1462-2920.2010.02198.x. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- Lohr J, Munn CB, Wilson WH. Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl Environ Microbiol. 2007;73:2976–2981. doi: 10.1128/AEM.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Jorgensen NOG, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl Environ Microbiol. 1996;62:1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TFF, Manire C, Borrowman K, Langer T, Ehrhart L, Breitbart M. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J Virol. 2009;83:2500–2509. doi: 10.1128/JVI.01946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor LM, Fuhrman JA. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–61. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes M, Coma R, Gili JM. Heterotrophic feeding by gorgonian corals with symbiotic zooxanthella. Limnol Oceanogr. 1998;43:1170–1179. doi: 10.4319/lo.1998.43.6.1170. [DOI] [Google Scholar]
- Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of viral metagenomes from Yellowstone hot springs. Appl Environ Microbiol. 2008;74:4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GW, Ives LD, Nagelkerken IA, Ritchie KB. Caribbean sea-fan mortalities. Nature. 1996;383:487. doi: 10.1038/383487a0. [DOI] [Google Scholar]
- Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159:349–357. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M (2010) Threatened corals provide underexplored microbial habitats. PLoS ONE 5(3): e9554. doi:10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed]
- Thingstad TF, Lignell R. A theoretical approach to the question of how trophic interactions control carbon demand, growth rate, abundance and diversity. Aquat Microb Ecol. 1997;13:19–27. doi: 10.3354/ame013019. [DOI] [Google Scholar]
- Thurber RLV, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, Edwards RA, Haynes M, Angly FE, Wegley L, Rohwer FL. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Toledo-Hernandez C, Bones-Gonzalez A, Ortiz-Vazquez OE, Sabat AM, Bayman P. Fungi in the sea fan Gorgonia ventalina: diversity and sampling strategies. Coral Reefs. 2007;26:725–730. doi: 10.1007/s00338-007-0252-8. [DOI] [Google Scholar]
- Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Francis I, Ryan K, Davy SK. Temperature induction of viruses in symbiotic dinoflagellates. Aquat Microb Ecol. 2001;25:99–102. doi: 10.3354/ame025099. [DOI] [Google Scholar]
- Wilson WH, Dale A, Davy JE, Davy SK. An enemy within? Observations of virus-like particles in reef corals. Coral Reefs. 2005;24:145–148. doi: 10.1007/s00338-004-0448-0. [DOI] [Google Scholar]


