Abstract
As a broad generalization, clinicians and laboratory personnel who use microfluidics-based automated or semi-automated instrumentation to perform biomedical assays on real-world samples are more pleased with the state of the assays than they are with the state of the front-end sample preparation. The end-to-end procedure requires one to collect, manipulate, prepare, and analyze the sample. The appeal of microfluidics for this procedure is partly based on its combination of small size and its ability to process very small liquid volumes, thus minimizing the use of possibly expensive reagents. However, real-world samples are often large and incompatible with the input port and the μm-scale channels of a microfluidic device, and very small liquid volumes can be inappropriate in analyzing low concentrations of target analytes. It can be a worthy challenge to take a raw sample, introduce it into a microfluidics-based system, and perform the sample preparation, which may include separation and concentration of the target analytes, so that one can benefit from the reagent-conserving small volumes and obtain the correct answer when finally implementing the assay of interest.
Keywords: Microfluidics, Biological assays, Sample preparation, Acoustics, Electrokinetics, Integration
Introduction—what is needed?
This article addresses instrumentation that is designed to perform biological assays (“bioassays”), and one overriding question in the design of any instrument that is to perform a bioassay is, simply put, “Who cares?” Who will use it and what actions will be taken, depending upon the results? These considerations drive everything “upstream” and greatly influence the selection of the sample(s), the processing of the sample, and the selection of assay(s) that are applied to the problem. Numerous of us who work with microfluidics-based systems have expressed optimism that some day we will have a self-powered, credit-card-sized, microfluidics-based, disposable cartridge that relatively untrained third parties can contact with a sample in some manner and, thereupon, the cartridge will transport and prepare the sample, mix it with reagents and execute other necessary sample processing steps, and then proceed to perform one or more identification-capable (ID-capable) multiplex biological assays, reporting its results for easy interpretation and action. We are not there, today, in general.
A useful example of a “simple,” single-target bioassay that nominally requires no sample preparation or user training is found in the very popular home pregnancy test (HPT) kits, which typically employ two monoclonal antibodies for human chorionic gonadotropin (hCG) in a lateral-flow, “sandwich” assay format. With this implementation, no formal instrumentation is required and the colorimetric readout is performed by eye. Bastian et al. have reviewed the use of such HPT Kits (Bastian et al. 1998) and cited studies that found nearly 24% to 50% false negatives, and “The main explanation for such high rate of false-negative results was difficulty in understanding the instruction in the HPT kits...,” Some information that is provided is:
The urine should be the first urine of the morning when hCG levels are highest [Urine becomes much more dilute after ingestion of liquids (tea, coffee, juice, water, etc.) and urine hCG concentrations may become too low to register as positive], but no particular sample preparation is typically employed. The antigen/antibody binding is particularly strong and confounding compounds/non-specific binding is normally not a problem. If a woman questions the result, she can wait until the next morning to repeat the measurement, normally with little in the way of negative consequence to a false positive or negative, while she waits.
This assay relies upon high-affinity, high-specificity antibodies to hCG and its subunits (Moyle et al. 1983), and whose concentrations have been well-studied, as function of age, in non-pregnant subjects (Alfthan et al. 1992; Odell and Griffin 1987; Snyder et al. 2005). Nonetheless, “... women may be obtaining their samples before the recommended number of days after their first missed menstrual period (usually 9 days)...,”(Bastian et al. 1998) which may be a major factor in the high false-negative rate.
Such lateral-flow strips that implement single-target “sandwich” assays using both mobile antibodies as well as immobilized antibodies comprise the most physically-robust, easy-to-use “detectors”.1 Nonetheless, even this very well-developed and “simple” manual implementation of an assay that only looks for one, well-known target and needs no sample preparation does, still, require that the user pay attention to instructions in order to attain reliable performance. One needs to carry forward this lesson in “end-to-end” system design, when creating a microfluidics-based system – consider whether there are anticipatable and likely “human errors” that may be associated with the difficulty of collecting a useful sample and introducing it into the input port.
Of course, designing and implementing a bioassay is vastly more difficult if one does not know what one is looking for (Rota et al. 2003), or even if one need look for an infectious agent at all, as was the case, when in 1984 followers of the Indian guru Bhagwan Shree Rajneesh spiked salad bars at ten restaurants with salmonella and sickened about 750 people (Torok et al. 1997). The same situation applied to the unintentional contamination of thousands of sandwiches with staphylococcus (Do Carmo et al. 2004).
Detecting small numbers of atoms or molecules in a sample
Thirty years ago, scientists first announced the ability to detect a single atom as it flew through a resonantly tuned laser beam (Dagenais and Mandel 1978; Hurst et al. 1977; Lewis et al. 1979). 10 years later, the ability to detect single phycoerythrin molecules (Nguyen et al. 1987) and individual, stained chromosomes (Gray et al. 1987) in a flow cytometer (Vandilla et al. 1969) was demonstrated. Also, inelastic tunneling spectroscopy detected, but did not identify, few impurity molecules in a tunnel junction (Sanchez et al. 1975).
For a given “s”, where s is defined as the product of sample volume multiplied by target concentration [which is, nominally, the average number of target molecules in a sample], Poisson-type statistics (Poisson 1837) quantify the probability that “n” targets will be present in the sample, assuming a random distribution in the original source.2 Then, the probability of actually having “n” targets in a sample that contains an average of s targets is given by
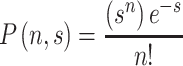 |
Thus, if s = 1, then the probability of finding no target is 37% [for n = 0, the probability P = 1/e = 0.37], so that, on average, even with the most accurate “single-molecule detection” you will not detect your targets 37% of the time, even if they are present in the larger sample! This assumes profound significance, when applying the small volume-handling capacity of microfluidics to biological assays at low, but physiologically relevant concentrations (Manz et al. 1990; Petersen et al. 1998; USFDA 2007).
Although not fully ID-capable for biological molecules, “giant” surface-enhanced-Raman-scattering [“SERS”] offers single-molecule detection that illustrates such Poisson statistical distributions (Kneipp et al. 1998). Figure 1 shows experimental data from SERS of adenine, with the calculated average sample sizes and the observed experimental distributions compared with theoretical distributions for s = 1.3 and 18 (estimated probe volume was 100 fL).
Fig. 1.
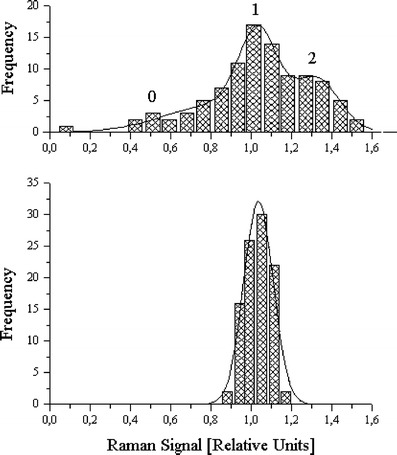
Reprinted figure with permission from K. Kneipp, H. Kneipp, V.B. Kartha, R. Manoharan, G. Deinum, I. Itzkan, R.R. Dasari, M.S. Feld, Physical Review E 57, R6281 (1998)
Nucleic-acid-based assays—very high information content
Since 1988, scientists have created highly specific genetic assays that worked down to the Poisson limit, such as the polymerase chain reaction(Lagally et al. 2001; Saiki et al. 1988), and these assays have been successfully applied to the detection of pathogens in difficult samples(Breitbart et al. 2003; Girones et al. 1993); microarray techniques have recently shown the capability to detect and identify respiratory viruses from clinical samples at physiologically relevant titers (Palacios et al. 2007; Wang et al. 2003, 2006). For blood-borne microbes, protein microarrays can work with diluted blood (Sundaresh et al. 2006), and, working with nasopharyngeal swab samples (McBride 2006), a semi-automated system has been demonstrated to detect conserved sequences of some known respiratory viruses. Note that in the demonstration of a powerful, multiplex bioassay for strain-specific identification of respiratory viruses (Wang et al. 2006), it was necessary to passage viruses through green monkey kidney cells as part of sample preparation, and in (Wang et al. 2003) it was necessary to scrape off the hybridized viral nucleic acid, manually, then clone and sequence it in order to identify this new pathogen.
Marriage of microfluidics with bioassays
Integrated microfluidic technologies promise to reduce costs and training requirements for high-performance, identification-capable bioanalysis, and efforts have been made for more than 20 years to create affordable, disposable plastic cartridges based on microfluidic technologies to be used with biological assays. See, e.g., Fig. 2 of a non-ID-capable, plastic microfluidics concept, based on the centrifugal action of a spinning compact disk (Kelton et al. 1981). See also Fig. 3, below, of a fully-ID-capable system that included a chamber for thermal cycling and that used a roller for the manual transport of sample and reagents (Schnipelsky et al. 1993).
Fig. 2.
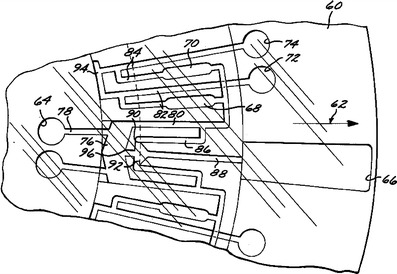
Taken from 1981 US Patent 4,284,602, this drawing illustrates the concept of using a spinning, multilayer “compact-disk” format to transport samples and reagents in microchannels from inner-radius locations, outwards
Fig. 3.
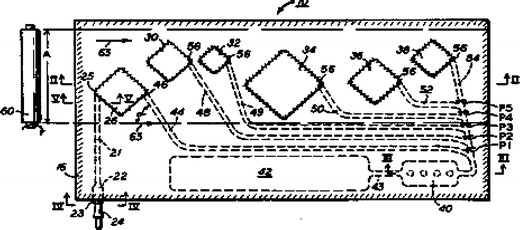
Taken from 1993 US Patent 5,229,297, this drawing illustrates the concept of a flexible, bubble-encapsulation approach for storage and transport of sample and reagents, with a roller to force fluids into and through microchannels
Microfluidics with nucleic-acid-based assays
Indeed, there have been encouraging results using microfluidic systems, once manual sample preparation was performed.
Bauemner and colleagues detected Dengue Virus RNA (Zaytseva et al. 2005) in a microfluidic biosensor.
The presence of SARS-coronavirus in nasopharyngeal swabs was confirmed, after using Qiagen RNAeasy kit to extract the nucleic acid (Zhou et al. 2004).
Lagally, Mathies, and colleagues used polymerase chain reaction (PCR) to detect methicillin resistance in Staphylococcus aureus, using a microfabricated, 200-nl PCR chamber, starting from suspensions of S. aureus grown in YPC media, diluted to 107 ml-1 in deionized water and added immediately to the PCR mix (Lagally et al. 2004). Detection and discrimination of strains was performed via fragment-length analysis with on-chip capillary-zone electrophoresis.
Quake and colleagues used 1,176 parallel, 6-nl PCR chambers to monitor and enumerate the genetic information of individual Treponema bacteria that they had extracted from termite hindguts (Ottesen et al. 2006).
Backhouse and co-workers demonstrated two-chip analysis of urine samples, using off-chip dilution, mixing with PCR master mix, etc, where the 1:100 overall dilution eliminated interference of inhibitors on the PCR-based assay (Kaigala et al. 2006). The presence of amplified product was determined on the second chip, which performed capillary-zone electrophoresis; the reaction mixture from the first chip was manually loaded into the second chip, with the addition of a sequencing ladder.
Microarray-based identification and enumeration of bacterial species in saliva has been demonstrated, after manual sample preparation (Starke et al. 2007), using 16S rRNA phylogenetics-based sequences; this non-amplification approach cannot be applied to detect viruses or plasmids.
Sample preparation in microfluidics
Initially, as can be seen in Figs. 4 and 5, below, showing the first state-of-the-art, ID-capable microtechnology-based instruments being used in the field (Belgrader et al. 1998b; Emanuel et al. 2003; Northrup et al. 1996), such high-performance, portable instruments based on genomic assays required skilled technical personnel, typically using gloves and micropipettors. Soon, however, microfluidics-based systems were being demonstrated for use in separating, purifying, and/or characterizing biological particles.
Fig. 4.

Dr. Phillip Belgrader, operating the first portable PCR instrument at Dugway Proving Grounds, 1997. See, also (Belgrader et al. 1998a; Northrup et al. 1996)
Fig. 5.

Dr. Michael Horseman, of Edgewood Chem-Bio Center, using the “Handheld Advanced Nucleic-Acid Analyzer”, 2001 (Emanuel et al. 2003; Northrup et al. 1996)
Gascoyne and co-workers diluted a suspension of malaria-infected, cultured erythrocytes into a buffer solution with conductivity = 55 mS/m and then successfully performed separations of the malaria-infected erythrocytes based on dielectrophoresis (Gascoyne et al. 2002; Gascoyne et al. 2004).
Lagally, Lee, and Soh (Lagally et al. 2005) used an integrated system for dielectrophoretic concentration and detection of cells, including 16S rRNA-based identification with a molecular beacon. The sample had been centrifuged, resuspended and diluted, prior to introduction to the microfluidic chip; flow rate was 0.1 ml/h.
Although some samples have too great a viscosity or debris/particle content to be directly compatible with microfabricated filters and microfluidics, Yobas and colleagues (Ji et al. 2008) worked with whole blood that they diluted 200X in phosphate-buffered saline and demonstrated the separation of red blood cells (RBCs) from white blood cells (WBCs), passing 60% or more of the RBCs and capturing 70% or more of the WBCs.
Working with simulated samples, consisting of aqueous suspensions of bacteria, Stachowiak, et al., also used microfabricated filters as the front end in a modular microfluidic system that employed a protein-based assay (Stachowiak et al. 2007).
Integrated/autonomous systems
The use of silica to capture, purify, and release nucleic acids for subsequent analysis has been known for more than a decade (Boom et al. 1993), and a microfabricated embodiment of this technique, using oxidized silicon pillars has been demonstrated (Christel et al. 1999). More recently, an integrated system has been reported (Raja et al. 2005) that employed these oxidized silicon pillars for automated extraction of nucleic acids and followed this step with real-time PCR.
There are two examples of macroscopic systems that perform automated sample collection from the environment at regular intervals, followed by automated sample preparation and multiplex, ID-capable assays (Greenfield et al. 2006; Langlois et al. 2003; McBride et al. 2003). Examples of integrated systems with microfluidic subcomponents that are ID-capable are rare—Yang, et al., starting with a suspension of killed Escherichia coli and using only minimal manual pipetting, demonstrated a stacked, plastic microfluidic system that used electrokinetic transport and trapping with strand-displacement amplification and hybridization to a microarray to detect the shiga-like toxin gene in E. coli (Heller et al. 2000; Yang et al. 2002). This system had macro tubing and valves.
Also, Grodzinski and co-workers, starting with 1 ml of citrated rabbit blood that they spiked with 106 ml-1 E. coli, used magnetic-bead-antibody-capture of the E. coli to purify and wash that target, then performed PCR and hybridization assay for readout in a single-target system of the E. coli (Liu et al. 2004).
Landers and co-workers created an integrated system and, starting from blood or nasal aspirate with externally added lysis buffer, used on-chip solid-phase extraction of nucleic acid, five on-chip, normally-closed elastomeric valves, along with an external syringe pump and pneumatic source for the on-chip valves, to perform on-chip lysis, extraction, PCR, and fragment detection via electrophoresis (Easley et al. 2006).
Where are we?
Today, an Internet search returns more than 100,000 hits for “single-molecule detection,” so...
Why hasn’t the combination of microfluidics with these single-molecule assays produced a portable, “while-you-wait” instrument to tell you if a microbe—virus, fungus, or bacterium is causing your sore throat? With nasopharyngeal samples, aside from the Poisson statistical limits that we discussed, above, one important part of the answer is the difficulty of sample handling/sample preparation.3
Recently, at the behest of Defense Advanced Research Projects Agency, my colleagues have made a survey of numerous labs across the USA that performed biodetection assays. As is discussed (Dougherty et al. 2007), although those who are responsible for analyzing clinical and environmental samples are basically satisfied with the performance of their reagents and assays, they would like to see improvements in the front-end procedures of sample handling and sample preparation. This echoes the previous major finding by an NRC committee on ID-capable instrumentation (Vitko Copyright 2005, ISBN 0-309-0957X), Finding 6-1:
Sample preparation, including sample handling, transport, and system integration, represents the single most important challenge to be faced in the production of a detect-to-warn system that performs an identification assay.
Returning to what is causing your sore throat and the task of detecting and identifying causative microbes, one important reason that microfluidic techniques have not become permanent capabilities in labs that perform ID-capable assays for respiratory pathogens is that clinical samples such as nasopharyngeal swabs have viscosity/polymeric constituents, along with trapped air and debris, that are not immediately processible within microfluidic channels. This problem was alluded to as the “world-to-chip” interface (Ramsey 1999). Also, the time-tested centrifuge is always named as something that clinical personnel would like to replace, but its replacement, if it exists, has not made the transition from research topic to laboratory workhorse.
Microfluidics and techniques for handling and preparing samples
Certainly, microfluidics have utilized acoustic and electrokinetic techniques such as electroosmosis, electrophoresis, and dielectrophoresis for the transport, purification, and concentration of analytes (Wong et al. 2004). Ultrasonics/acoustics has effectively manipulated eukaryotic cells (Hawkes et al. 2002; Petersson et al. 2005); dielectrophoresis has separated bacteria from other components (Chiou et al. 2005; Pethig and Markx 1997; Yang et al. 2002); optoelectronic tweezers have manipulated particles (Chiou et al. 2005); isotachophoresis has demonstrated million-fold concentration under some circumstances (Jung et al. 2006). Thus, one can easily envision a series of microfluidics-based “virtual filters” through which particles are separated and concentrated, according to size: ultrasonics work well with cells that are larger than 2 μm, dielectrophoresis can trap bacteria and other particles that are small enough to pass through an acoustic separator and are between 1 and 2 μm in scale, and temperature gradient focusing (Shackman et al. 2007), isotachophoresis (Jung et al. 2006), and isoelectric focusing (Poitevin et al. 2007) may provide very fine separation and/or concentration of sub-μm constituents. For microfluidics, the promise is still very much there.
Nonetheless, why has it not progressed further? It is easy to list some reasons:
So far, the market for ID-capable biomedical microdevices has been relatively small, in general, so the sample-preparation portion of that has languished. This is not to mention that solving the world-to-chip interface has been difficult.
Large-scale/high-throughput sample preparation is typically performed in the pharmaceutical industry using microtiter plates and robotic systems. Microfluidics-based systems face stiff technical and cost competition as well as the “inertia” of the established technology and supporting infrastructure.
Microfluidic systems can be fabricated from a wide variety of materials, and a technique such as acoustic focusing can work very well in glass or silicon, but less well in hard plastics and not well at all in soft materials such as silicone rubber [e.g. PDMS]. Integration of disparate materials in a cost-effective, robust, and leak-free manner is a difficult task, even with off-chip pumps, valves, etc.
Medical point-of-care applications can be very specific, [see, for example, blood analyses such as Blood Gases, Electrolytes, Chemistries, Coagulation, Hematology, Glucose, and Cardiac Markers (cTnI) performed on the i-STAT®1 [http://www.abbottpointofcare.com/istat/www/products/analyzers.htm], and solving the problem of sample collection and preparation for one application may not position you to do the same for the next application you address.
Take-away message—the big picture for little instruments
Although advances are being made by scientists and engineers using Microtechnology and Nanotechnology for single-molecule detection, I urge the investigators who are working in these fields to devote time to solving the front-end problems of sample collection, sample handling, and sample preparation. Along these lines, I have found the following questions to stimulate valuable discussions for detection systems:
What overall problem are you addressing?
What scenarios are applicable and under what Concept of Operation do your assay and instrument fit in?
What is the state-of-the-art for this scenario and what limits it?
What are possible improvements and why did you choose the one you did?
What level of end-to-end performance is needed and how will you measure “success”?
I believe that no one better understands the strengths and limitations of the latest instruments than those who are creating them and, therefore, no one is better positioned to evaluate what samples are needed and how these samples need to be prepared in order to maximize their performance. When progress is made in the end-to-end process, we all “win.”
Acknowledgement
The author wishes to thank his colleagues, at LLNL and elsewhere, who have encouraged and supported him in pursuing the “mission” of sample preparation for fieldable systems. The author also wishes to thank Reviewer#1 for suggesting valuable revisions to this manuscript.
Footnotes
One can drop a LFA strip onto a hard surface and still have a good chance that it will work as well as before the drop.
If the target molecules are not uniformly/randomly distributed, then the Poisson statistics may not apply.
Detecting and identifying pathogenic microbes in ground beef or on spinach is also a challenge, due to the combination of the low dose threshold [estimated by the US FDA to be 10 E. Coli O157:H7 bacteria USFDA, http://www.cfsan.fda.gov/~mow/chap15.html (2007). or 1 mg of SEB] with the large quantities of food being consumed. [See, e.g., thousands poisoned from eating infected sandwiches L.S. Do Carmo, C. Cummings, V.R. Linardi, R.S. Dias, J.M. De Souza, M.J. De Sena, D.A. Dos Santos, J.W. Shupp, R.K. Pereira, M. Jett, Foodborne pathogens and disease 1(4), 241 (2004)].
This work performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
References
- Alfthan H., Haglund C., Dabek J., Stenman U.H. Clin Chem. 1992;38(10):1981. [PubMed] [Google Scholar]
- Bastian L.A., Nanda K., Hasselblad V., Simel D.L. Arch Fam Med. 1998;7(5):465. doi: 10.1001/archfami.7.5.465. [DOI] [PubMed] [Google Scholar]
- Belgrader P., Benett W., Hadley D., Long G., Mariella R., Jr., Milanovich F., Nasarabadi S., Nelson W., Richards J., Stratton P. Clin Chem. 1998;44(10):2191. [PubMed] [Google Scholar]
- Belgrader P., Smith J.K., Weedn V.W., Northrup M.A. J Foren Sci. 1998;43(2):315. [PubMed] [Google Scholar]
- Boom W.R., Adriaanse H.M.A., Kievits T., Lens P.F. Process for isolating nucleic acid, US5234809A. Amsterdam: Azko Nobel; 1993. [Google Scholar]
- Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P., Rohwer F. J Bacteriol. 2003;185(20):6220. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou P.Y., Ohta A.T., Wu M.C. Nature. 2005;436(7049):370. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- Christel L.A., Petersen K., McMillan W., Northrup M.A. J Biomech Eng. 1999;121(1):22. doi: 10.1115/1.2798037. [DOI] [PubMed] [Google Scholar]
- Dagenais M., Mandel L. Phys Rev A. 1978;18(5):2217. doi: 10.1103/PhysRevA.18.2217. [DOI] [Google Scholar]
- Do Carmo L.S., Cummings C., Linardi V.R., Dias R.S., De Souza J.M., De Sena M.J., Dos Santos D.A., Shupp J.W., Pereira R.K., Jett M. Foodborne Path Dis. 2004;1(4):241. doi: 10.1089/fpd.2004.1.241. [DOI] [PubMed] [Google Scholar]
- G.M. Dougherty, D.S. Clague, R.R. Miles, In Saito, T.T., Lehrfeld, D., DeWeert, M.J. (Eds.), Optics and Photonics in Global Homeland Security III. Proc SPIE. 6540, 54016 (2007)
- Easley C.J., Karlinsey J.M., Bienvenue J.M., Legendre L.A., Roper M.G., Feldman S.H., Hughes M.A., Hewlett E.L., Merkel T.J., Ferrance J.P., Landers J.P. Proc Nat Acad Sci U S A. 2006;103(51):19272. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel P.A., Bell R., Dang J.L., McClanahan R., David J.C., Burgess R.J., Thompson J., Collins L., Hadfield T. J Clin Microbiol. 2003;41(2):689. doi: 10.1128/JCM.41.2.689-693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P., Mahidol C., Ruchirawat M., Satayavivad J., Watcharasit P., Becker F.F. Lab on a chip. 2002;2(2):70. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P., Satayavivad J., Ruchirawat M. Acta Tropica. 2004;89(3):357. doi: 10.1016/j.actatropica.2003.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girones R., Allard A., Wadell G., Jofre J. Water Sci Technol. 1993;27(3–4):235. [Google Scholar]
- Gray J.W., Dean P.N., Fuscoe J.C., Peters D.C., Trask B.J., Vandenengh G.J., Vandilla M.A. Science. 1987;238(4825):323. doi: 10.1126/science.2443974. [DOI] [PubMed] [Google Scholar]
- Greenfield D.I., Marin R., Jensen S., Massion E., Roman B., Feldman J., Scholin C.A. Limnol Oceanog -Methods. 2006;4:426. [Google Scholar]
- Hawkes J.J., Coakley W.T., Gröschl M., Benes E., Armstrong S., Tasker P.J., Nowotny H. J Acoust Soc Am. 2002;111(3):1259. doi: 10.1121/1.1448341. [DOI] [PubMed] [Google Scholar]
- Heller M.J., Forster A.H., Tu E. Electrophoresis. 2000;21(1):157. doi: 10.1002/(SICI)1522-2683(20000101)21:1<157::AID-ELPS157>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Hurst G.S., Nayfeh M.H., Young J.P. Appl Phys Lett. 1977;30(5):229. doi: 10.1063/1.89360. [DOI] [Google Scholar]
- Ji H.M., Samper V., Chen Y., Heng C.K., Lim T.M., Yobas L. Biomed Microdevices. 2008;10(2):251. doi: 10.1007/s10544-007-9131-x. [DOI] [PubMed] [Google Scholar]
- Jung B., Bharadwaj R., Santiago J.G. Anal Chem. 2006;78(7):2319. doi: 10.1021/ac051659w. [DOI] [PubMed] [Google Scholar]
- Kaigala G.V., Huskins R.J., Preiksaitis J., Pang X.L., Pilarski L.M., Backhouse C.J. Electrophoresis. 2006;27(19):3753. doi: 10.1002/elps.200600061. [DOI] [PubMed] [Google Scholar]
- Kelton A.A., Waters W.P., Shrunk D.G., Bell M.L. Integrated fluid manipulator, US4284602A. Newport Beach: Immutron; 1981. [Google Scholar]
- Kneipp K., Kneipp H., Kartha V.B., Manoharan R., Deinum G., Itzkan I., Dasari R.R., Feld M.S. Phys Rev E. 1998;57(6):R6281. doi: 10.1103/PhysRevE.57.R6281. [DOI] [Google Scholar]
- Lagally E.T., Medintz I., Mathies R.A. Anal Chem. 2001;73(3):565. doi: 10.1021/ac001026b. [DOI] [PubMed] [Google Scholar]
- Lagally E.T., Scherer J.R., Blazej R.G., Toriello N.M., Diep B.A., Ramchandani M., Sensabaugh G.F., Riley L.W., Mathies R.A. Anal Chem. 2004;76(11):3162. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- Lagally E.T., Lee S.H., Soh H.T. Lab on a chip. 2005;5(10):1053. doi: 10.1039/b505915a. [DOI] [PubMed] [Google Scholar]
- R.G. Langlois, F.P. Milanovich, B.W. Colston, S.B. Brown, D.A. Masquelier, R.P. Mariella, K. Venkateswaran, System for autonomous monitoring of bioagents, EP1576353 (US20040038385A1) (2003)
- Lewis D.A., Tonn J.F., Kaufman S.L., Greenlees G.W. Phys Rev A. 1979;19(4):1580. doi: 10.1103/PhysRevA.19.1580. [DOI] [Google Scholar]
- Liu R.H., Yang J.N., Lenigk R., Bonanno J., Grodzinski P. Anal Chem. 2004;76(7):1824. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- Manz A., Graber N., Widmer H.M. Sens Actuators B-Chem. 1990;1(1–6):244. doi: 10.1016/0925-4005(90)80209-I. [DOI] [Google Scholar]
- M.T. McBride, LLNL Sci Technol Rev. 4–9 (2006)
- McBride M.T., Masquelier D., Hindson B.J., Makarewicz A.J., Brown S., Burris K., Metz T., Langlois R.G., Tsang K.W., Bryan R., Anderson D.A., Venkateswaran K.S., Milanovich F.P., Colston Jr B.W. Anal Chem. 2003;75(20):5293. doi: 10.1021/ac034722v. [DOI] [PubMed] [Google Scholar]
- Moyle W.R., Lin C., Corson R.L., Ehrlich P.H. Mol Immunol. 1983;20(4):439. doi: 10.1016/0161-5890(83)90025-1. [DOI] [PubMed] [Google Scholar]
- Nguyen D.C., Keller R.A., Jett J.H., Martin J.C. Anal Chem. 1987;59(17):2158. doi: 10.1021/ac00144a032. [DOI] [PubMed] [Google Scholar]
- Northrup M.A., Mariella J.R.P., Carrano A.V., Balch J.W. Silicon-based sleeve devices for chemical reactions, US5589136A. Oakland: Regents of the University of California; 1996. [Google Scholar]
- Odell W.D., Griffin J. New Eng J Med. 1987;317(27):1688. doi: 10.1056/NEJM198712313172702. [DOI] [PubMed] [Google Scholar]
- Ottesen E.A., Hong J.W., Quake S.R., Leadbetter J.R. Science. 2006;314(5804):1464. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Palacios G., Quan P.-L., Jabado O., Conlan S., Hirschberg D., Liu Y., et al. Emerg Infect Dis. 2007;13(1):73. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K.E., McMillan W.A., Kovacs T.A., Northrup M.A., Christel L.A., Pourahmadi F. Biomed Microdev. 1998;1(1):71. doi: 10.1023/A:1009986407026. [DOI] [Google Scholar]
- Petersson F., Nilsson A., Holm C., Jonsson H., Laurell T. Lab on a chip. 2005;5(1):20. doi: 10.1039/b405748c. [DOI] [PubMed] [Google Scholar]
- Pethig R., Markx G.H. Trends Biotechnol. 1997;15(10):426. doi: 10.1016/S0167-7799(97)01096-2. [DOI] [PubMed] [Google Scholar]
- Poisson S.D. Treatise. Paris: Ecole Polytechnique; 1837. [Google Scholar]
- Poitevin M., Morin A., Busnel J.M., Descroix S., Hennion M.C., Peltre G. J Chromatogr A. 2007;1155(2):230–236. doi: 10.1016/j.chroma.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Raja S., Ching J., Xi L.Q., Hughes S.J., Chang R., Wong W., McMillan W., Gooding W.E., McCarty K.S., Chestney M., Luketich J.D., Godfrey T.E. Clin Chem. 2005;51(5):882. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- Ramsey J.M. Nature Biotechnol. 1999;17(11):1061. doi: 10.1038/15044. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300(5624):1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saiki R.K., Gelfand D.H., Stoffel S., Scharf S.J., Higuchi R., Horn G.T., Mullis K.B., Erlich H.A. Science. 1988;239(4839):487. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Davis C.F., Jr., Elchinger G.M., Javan A. 29th Annual Symposium on Frequency control. 1975;1975:328–329. [Google Scholar]
- Schnipelsky P.N., Seaberg L.J., Hinckley C.C., Wellman J.A., Donish W.H., Findlay J.B. Containment cuvette for PCR and method of use, US5229297A. Rochester: Eastman Kodak; 1993. [Google Scholar]
- Shackman J.G., Munson M.S., Ross D. Anal Bioanal Chem. 2007;387(1):155. doi: 10.1007/s00216-006-0913-4. [DOI] [PubMed] [Google Scholar]
- Snyder J.A., Haymond S., Parvin C.A., Gronowski A.M., Grenache D.G. Clin Chem. 2005;51(10):1830. doi: 10.1373/clinchem.2005.053595. [DOI] [PubMed] [Google Scholar]
- Stachowiak J.C., Shugard E.E., Mosier B.P., Renzi R.F., Caton P.F., Ferko S.M., Van de Vreugde J.L., Yee D.D., Haroldsen B.L., VanderNoot V.A. Anal Chem. 2007;79(15):5763. doi: 10.1021/ac070567z. [DOI] [PubMed] [Google Scholar]
- Starke E.M., Smoot J.C., Wu J.-H., Liu W.-T., Chandler D., Stahl D.A. Ann NY Acad Sci. 2007;1098(1):345. doi: 10.1196/annals.1384.007. [DOI] [PubMed] [Google Scholar]
- Sundaresh S., Doolan D.L., Hirst S., Mu Y., Unal B., Davies D.H., Felgner P.L., Baldi P. Bioinformatics (Oxford). 2006;22(14):1760. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- Torok T.J., Tauxe R.V., Wise R.P., Livengood J.R., Sokolow R., Mauvais S., Birkness K.A., Skeels M.R., Horan J.M., Foster L.R. JAMA. 1997;278(5):389. doi: 10.1001/jama.278.5.389. [DOI] [PubMed] [Google Scholar]
- USFDA, Foodborne pathogenic microorganisms and natural toxins handbook, Accessed at http://www.cfsan.fda.gov~mow/chap15.html/ (2007)
- Vandilla M.A., Trujillo T.T., Mullaney P.F., Coulter J.R. Science. 1969;163(3872):1213. doi: 10.1126/science.163.3872.1213. [DOI] [PubMed] [Google Scholar]
- Vitko J., Jr. In: Committee on Materials and Manufacturing Processes for Advanced Sensors, National Research Council. Design, B.o.M.a.E., editor. Washington, D.C.: National Academies; 2001. [Google Scholar]
- Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Latreille J.P., Wilson R.K., Ganem D., DeRisi J.L. Plos Biology. 2003;1(2):257. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B.C., Stenger D.A. Emerg Infect Dis. 2006;12(4):638. doi: 10.3201/eid1204.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.K., Wang T.H., Deval J.H., Ho C.M. IEEE-ASME Transact Mechatron. 2004;9(2):366. doi: 10.1109/TMECH.2004.828659. [DOI] [Google Scholar]
- Yang J.M., Bell J., Huang Y., Tirado M., Thomas D., Forster A.H., Haigis R.W., Swanson P.D., Wallace R.B., Martinsons B., Krihak M. Biosens Bioelectron. 2002;17(6–7):605. doi: 10.1016/S0956-5663(02)00023-4. [DOI] [PubMed] [Google Scholar]
- Zaytseva N.V., Montagna R.A., Baeumner A.J. Anal Chem. 2005;77(23):7520. doi: 10.1021/ac0509206. [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu D., Zhong R., Dai Z., Wu D., Wang H., Du Y., Xia Z., Zhang L., Mei X., Lin B. Electrophoresis. 2004;25(17):3032. doi: 10.1002/elps.200305966. [DOI] [PMC free article] [PubMed] [Google Scholar]


