Abstract
Pathogenic free-living amoebae (FLA), such as Naegleria fowleri, Balamuthia mandrillaris and Acanthamoeba species isolated from aquatic environments have been implicated in central nervous system, eye and skin human infections. They also allow the survival, growth and transmission of bacteria such as Legionella, Mycobacteria and Vibrio species in water systems. The purpose of this study was to investigate the co-occurrence of potentially pathogenic FLA and their associated bacteria in hospital water networks in Johannesburg, South Africa. A total of 178 water (n = 95) and swab (n = 83) samples were collected from two hospital water distribution systems. FLA were isolated using the amoebal enrichment technique and identified using PCR and 18S rDNA sequencing. Amoebae potentially containing intra-amoebal bacteria were lysed and cultured on blood agar plates. Bacterial isolates were characterized using the VITEK®2 compact System. Free-living amoebae were isolated from 77 (43.3 %) of the samples. Using microscopy, PCR and 18S rRNA sequencing, Acanthamoeba spp. (T3 and T20 genotypes), Vermamoeba vermiformis and Naegleria gruberi specie were identified. The Acanthamoeba T3 and T20 genotypes have been implicated in eye and central nervous system infections. The most commonly detected bacterial species were Serratia marcescens, Stenotrophomonas maltophilia, Delftia acidovorans, Sphingomonas paucimobilis and Comamonas testosteroni. These nosocomial pathogenic bacteria are associated with systematic blood, respiratory tract, the urinary tract, surgical wounds and soft tissues infections. The detection of FLA and their associated opportunistic bacteria in the hospital water systems point out to a potential health risk to immune-compromised individuals.
Electronic supplementary material
The online version of this article (doi:10.1007/s00436-016-5271-3) contains supplementary material, which is available to authorized users.
Keywords: Amoebal enrichment, Acanthamoeba spp., Vermamoeba vermiformis, Serretia marcescens
Introduction
Hospital water supplies often contain waterborne pathogens such as bacteria, viruses, fungi, algae and protozoa, which can be a source of healthcare-associated infections (HAIs) (Anaissie et al. 2002; Stojek et al. 2008; Laganà et al. 2014). Among the protozoa, free-living amoebae (FLA) exist in high numbers in aquatic environments where they play a useful role as predators of micro-organisms, contributing to nutrient recycling by assimilating nutrients such as carbon, nitrogen and phosphate and releasing them into the environment by excretion (Taylor 1982; Rodriguez-Zaragoza 1994; Siddiqui and Khan 2012). Although mostly non-pathogenic, some FLA such as Acanthamoeba, Naegleria fowleri, Balamuthia mandrillaris and Sappinia pedata have been implicated in opportunistic and non-opportunistic eye, skin and central nervous system human infections (Visvesvara et al. 2007; Visvesvara 2013; Fuerst et al. 2015).
In addition to causing infections, FLA can also interact with and act as reservoirs and transmitters of waterborne pathogens. Organisms mostly associated with HAIs such as Legionella spp., Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus, Acinetobacter spp., Serratia spp., Klebsiella spp. and Aspergillus spp. have been reported to coexist with FLA in the hospital environment (Thomas et al. 2006; Cateau et al. 2014; Laganà et al. 2014). These organisms can infect and survive the exposure to oxygen radicals, the acidic environment and the lysosomal enzymes in the phagosomes of FLA. In addition to survival, some bacteria can multiply inside FLA (Greub and Raoul 2004; Lone et al. 2009; Siddiqui and Khan 2012). Organisms in this category are collectively known as “endocytobionts” (Scheid 2014). FLA may upregulate virulent genes of endocytobionts resulting in increased infectivity for human macrophages in vitro (Schmitz-Esser et al. 2008; Siddiqui and Khan 2012). Furthermore, the interaction of amoebae with pathogenic endocytobionts can decrease the susceptibility of these pathogens to biocides mainly used to treat water in water systems (Miltner and Bermudez 2000; Coulon et al. 2010). Regular monitoring of FLA and associated endocytobionts in hospital and other health care institutions is thus essential to manage and prevent possible exposure to patients, staff and members of the public who visit these institutions. In South Africa, contamination of hospital water supplies with waterborne pathogens has largely been overlooked. To our knowledge, this is the first study to investigate the co-occurrence of potentially pathogenic FLA and their associated bacteria in two selected public hospital water networks in South Africa.
Methods
Sampling
From February to December 2014, a total of 178 samples, (95 water and 83 swab), were collected from the water distribution systems of two public hospitals in Johannesburg (South Africa): 30 from hospital A and 148 from hospital B. Samples were collected from the municipal water inlets, theatres, intensive care units (ICU), endoscopy units, renal units, neonatal ward, neonatal ICU milk rooms, diarrhea ward and sterilization units of the two hospitals. Biofilm samples were collected by swabbing inside the taps prior to opening them. Water samples were collected after running the taps for 1–2 min in 1-L sterile sampling bottles containing 5 mg/L sodium thiosulfate (Merck, SA) to neutralize the chlorine disinfectant and oxidizing biocides which may be present at the time of sampling. At each sampling site, water temperature and pH were measured with a portable COMBO TESTER® (Hanna, SA) according to the manufacturer’s instructions. Residual chlorine was measured using a chlorine photometer (Hanna, SA) according to the manufacturer’s instructions. The samples were collected and transported in cooler boxes to keep them at ambient temperature to the laboratory and processed on the day of receipt.
Isolation of FLA
Five hundred millilitres of water sample was concentrated by filtration using a nitrocellulose membrane (Millipore, SA) with a pore size of 0.45 μm. Swabs were vortexed at maximum speed for 30 s in 10 mL Page’s amoebal saline buffer (PAS) in individual sterile tubes, and the suspension was also concentrated by membrane filtration like the water samples. Each filter membrane was placed upside down onto a non-nutrient agar (NNA) plate overlaid with a suspension of heat-killed Escherichia coli (E. coli) ATCC 25922 (E. coli; 100 μL for each plate). The plates were incubated aerobically at 32 °C and examined daily for 3 weeks under a light microscope (Olympus, Japan) with ×10 objective for appearance of amoebal trophozoites and/or cysts. Plates were recorded as negative if no amoebae were observed after 3 weeks. Plates containing amoebae were sub-cultured by cutting small agar plugs, placing them upside down onto fresh NNA-E. coli plates and incubating as before. Sub-culturing was done three times to purify amoebae cells. Once purified, amoebae cells were harvested by gently scraping the agar surface and re-suspending in 1 mL PAS. To further remove extracellular bacteria and debris, the suspension was centrifuged three times at 1000×g for 20 min. The washed pellet was re-suspended in 1 mL PAS. The suspension was inoculated into 24-well microtitre plate wells (Nunc, USA) and incubated at 32 °C. The plates were checked daily under an inverted microscope (Leica, Germany), with a ×40 objective, for the morphological appearance of FLA. Transmission electron microscopy (TEM) was used to screen for the presence of intracellular bacteria
Transmission electron microscopy
Each non-nutrient agar plate with amoebal trophozoites was flooded with fixative and left overnight. The fixative contained 2.5 % glutaraldehyde and 2 % formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.15), osmotically adjusted by the addition of 0.01 M CaCl2, 0.01 M MgCl2 and 0.09 M sucrose. The agar surface was then scraped, and for subsequent steps in the protocol, the sample was pelleted by gentle centrifugation (300×g for 20 min). Buffer rinses (30 min each) and post-fixation with 1 % osmium tetroxide for 1 h preceded graded ethanol dehydration and embedding in a low viscosity resin. Sections of 70 nm were cut on a Leica EM UC6 ultramicrotome, placed on Formvar®-coated slot grids, double stained with uranyl acetate and lead citrate, and viewed on an FEI BioTwin Spirit transmission electron microscope at 80 kV (FEI, USA). Images were captured with an Olympus Quemesa CCD camera (Olympus, Germany).
Molecular detection of FLA
DNA extraction and amplification
Amoebal DNA was extracted from 200 μL of the prepared amoebae suspension (from each of the amoebae positive plates) using the QIAamp DNA Blood Mini Kit (Qiagen, Germany), according to the manufacturer’s protocol. The nucleic acid was eluted in 100 μL elution buffer into a 1.5-mL micro-centrifuge tube and stored at −20 °C for subsequent analysis by PCR, agarose gel electrophoresis and sequence analysis. An 18S rDNA PCR was performed using the primers Ami6F1 (5′-CCA GCT CCA ATA GCG TAT ATT-3′), Ami6F2 (5′-CCA GCT CCA AGA GTG TAT ATT-3′) and Ami9R (5′-GTT GAG TCG AAT TAA GCC GC-3′) which amplifies approximately 700 bp of the 18 rDNA gene (Table 1) at a concentration of 0.5 μM (Thomas et al. 2006) with Takyon PCR Master Mix (Eurogentec, Belgium). After the first step consisting of 94 °C for 5 min, 40 cycles of amplification were performed by using denaturation at 94 °C for 1 min, annealing at 55 °C for 30s and elongation at 72 °C for 2 min and a final cycle at 72 °C for 10 min.
Table 1.
Primers used for amplification
| Organism | Target gene | Primer | Sequence | Bp | Reference |
|---|---|---|---|---|---|
| FLA | 18S rDNA |
Ami6F1 Ami6F2 Ami9R |
5′CCAGCTCCAATAGCGTATATT3′ 5′CCAGCTCCAAGAGTGTATATT3′ 5′GTTGAGTCGAATTAAGCCGC3′ |
700 | Thomas et al. 2006 |
| Acanthamoeba spp. | 18S rDNA |
JDP1 JDP2 |
5′GGCCCAGATCGTTTACCGTGAA3′ 5′TCTCACAAGCTGTAGGGGAGTCA3′ |
500 | Schroeder et al. 2001 |
Bp base pairs
DNA sequencing
Sequencing reactions were then performed with each primer. Sample purification was performed by using the Genelute Kit (Sigma-Aldrich, USA) following the manufacturer’s protocol. Efficiency of PCR has been confirmed by agarose gel electrophoresis with 8 μL of the PCR product on 2 % gel (data not shown). Genome content of the positive samples has been determined by photometric method (Qubit 2.0 Fluorometer, Life Technologies, USA) and subsequently sequenced with a 3130xl Genetic Analyzer (Applied Biosystems, USA). In order to determine Acanthamoeba genotype, follow-up PCR was performed with the primers set JDP1 (5′-GGC CCA GAT CGT TTA CCG TGA A-3′) and JDP2 (5′-TCT CAC AAG CTG CTA GGG GAG TCA-3′) which amplifies approximately 500 bp of the 18 rRNA gene (Table 1) (Schroeder et al. 2001). Cycling conditions were as follows: 95 °C for 5 min for the initial denaturation step, followed by 35 cycles of 15 s at 95 °C for denaturation, 15 s at 62 °C for annealing, 3 s at 72 °C for extension and a final extension at 72 °C for 10 min. PCR products of Acanthamoeba were confirmed like mentioned before via gel electrophoresis on a 2 % agarose gel. Phylogenetic construction produced gene trees by using neighbour-joining distance trees with a generation of 1,500 bootstrapped replicates. In order to allow BLAST searching and alignment with the MEGA 6.06 (Mega Software, USA) software, the 18S rDNA gene sequences were assigned to the GenBank database at the National Center for Biotechnical Information (NCBI). Isolates which have not been identified thus far were deposited in the GenBank under accession numbers KT18374-KT183626. Obtained sequences were aligned with sequences of Acanthamoeba genotypes T1–T20 (Corsaro et al. 2015).
Isolation and identification of amoebae-associated bacteria
Eight hundred microliters of the amoebae suspension from the microtitre plate was passed three times through a 27-gauge needle to lyse the amoebae and release any potential intracellular bacteria. Hundred microliters of the lysed amoebae suspension was then inoculated onto blood agar (NHLS, SA) and incubated at 37 °C overnight. Representative colonies were inoculated onto fresh blood agar plates and incubated as above. Gram stains were performed on the colonies to group them as Gram positive or negative. Pure colonies were placed in a tube containing 3 mL saline (BioMérieux, Inc.), and the density was monitored with the VITEK®2 DensiCHEK™. Bacterial suspensions not within the appropriate zone were adjusted to 0.5–0.63 McFarland standard. Once the correct density was achieved, the appropriate VITEK®2 Identification Card was selected and inserted into the tube containing the suspension. Thus, the VITEK®2 Identification Card for Gram-positive organisms was placed in the tube containing the Gram-positive bacterial suspension, and the VITEK®2 Identification Card for Gram-negative organisms was placed in the tube containing Gram negative bacterial suspension. Thereafter, the instructions on data entry and how to load the cassette into the instrument were followed as directed by the manual and instrument. After insertion of the cassettes into the instrument, the cassettes were incubated for 18–24 h, after which the organisms were identified using the VITEK®2 compact System software.
Statistical analysis
Statistical analysis was used to determine the differences in the isolation of amoebae from biofilm and water samples in hospitals A and B. The collected data were analyzed with SPSS® version 20 (SPSS Inc., USA), using crossing tables and chi-square test (asymptotic significance, two-tailed). Statistical significance was set at p < 0.05. The Pearson chi-square test was used to test for association between categorical variables. The interpretation was performed at a 95 % confidence limit.
Results
Physicochemical parameters
The water temperature of the two hospitals at the time of sampling ranged between 18.3 and 30.8 °C (mean 22.6 °C) at hospital A and 20.0–25.7 °C (mean 22.6 °C) at hospital B. The pH of the water ranged between 3.8 and 8.5 (mean 7.8); 7.6–8.5 (mean 7.8) at hospital A and 3.8–8.1 (mean 7.6) at hospital B. The pH of 3.8 was recorded from one sample collected at the renal dialysis unit of hospital A after treatment by reverse osmosis. The residual chlorine levels for the hospitals ranged between 0.01 and 0.34 mg/L (mean 0.28 mg/L) at the time of sampling. The chlorine levels of the individual hospitals were 0.06–0.33 mg/L (mean 0.23 mg/L) for hospital A and 0.01–0.34 mg/L (mean 0.20 mg/L) for hospital B.
Isolation of FLA
A total of 77/178 (43.3 %) of the samples collected at the two hospitals contained FLA (Table 2). There was no significant difference between the prevalence of FLA in water and biofilm samples (collected by swabbing inside the opening of taps) (P > 0.05), with 31 (32.6 %) of the FLA isolated from the water and 46 (55.4 %) from the biofilm after amoebal enrichment. Free-living amoebae were observed in 15 (50.0 %) of the samples, 7 (38.8 %) from the water and 8 (66.6 %) from the biofilm samples (P = 0.14) in hospital A. In hospital B, FLA was isolated from 62 (41.9 %) of the samples, 24 (31.0 %) from the water and 38 (53.5 %) from the biofilm (P = 0.25) samples. Amoebae were present in samples from all sampling sites, except from the municipal water inlet of hospital B. Based on morphology, three of the positive samples from hospital A contained clearly distinguishable Vermamoeba trophozoites. The FLA present in the other 12 samples could not be distinguished morphologically and were placed in the category “other FLA”. Vermamoeba species were observed in just over a half the (56.0 %), “other FLA” in 29 % and Acanthamoeba species in 15.0 % of hospital A samples. The TEM of isolated FLA revealed the occurrence of potential intracellular bacteria (Fig. 1) whose identities were determined by VITEK®2 analysis.
Table 2.
FLA isolated in water and biofilm samples in hospitals A and B
| Water + (%) | Biofilm + (%) | FLA + | |
|---|---|---|---|
| Area | Hospital A | ||
| Municipal water inlet | 1 (100) | 0 (0.0) | 1 |
| Neonatal ward | 3 (37.5) | 5 (100) | 8 |
| Neonatal ICU | 1 (33.3) | 2 (50.0) | 3 |
| Diarrhoea ward | 2 (50.0) | 1 (50.0) | 3 |
| Milk kitchen | 0 (0.0) | ND | 0 |
| Subtotal | 7 (38.8 %) | 8 (66.6 %) | 15 |
| Hospital B | |||
| Municipal water inlet | 0 (0.0) | 0 (0.0) | 0 |
| Renal unit | 1 (16.7) | ND | 1 |
| ICU | 10 (18.6) | 14 (41.1) | 24 |
| Theatre complex | 10 (45.5) | 11 (50.1) | 21 |
| TSSU | 1 (50.0) | 2 (100) | 3 |
| CSSU | 1 (50.0) | 2 (100) | 3 |
| Endoscopy/bronchoscopy | 1 (20.0) | 6 (100) | 7 |
| Milk kitchen | 0 (0.0) | 3 (50.0) | 3 |
| Total | 31 (32.6) | 46 (55.4) | 77 |
NB: ND not done
Fig. 1.
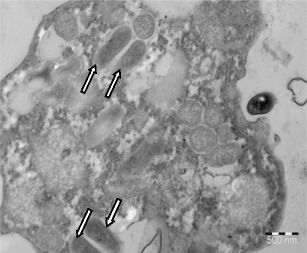
Transmission electron image of an amoeba with intracellular bacteria (arrows), scale bar 500 nm
Molecular identification of FLA
Fifteen amoebae-positive samples from hospital A were analyzed using PCR and DNA sequencing. Three culture plates were overgrown with fungi and were discarded. From the 12 amoebae-positive samples, a total of eight Vermamoeba vermiformis isolates detected, GenBank accession numbers: KT385808, KT385810, KT385815, KT38582 (from the neonatal ward); KT385807, KT385809 (from the neonatal ICU) and KT385811, KT385816 (from gastrointestinal ward). The other four Acanthamoeba spp. isolates were detected from the neonatal ward (KT385816, KT385819), neonatal ICU (KT385817) and gastrointestinal ward (KT385814). A total of 62 amoebae positive samples from hospital B were analyzed using PCR and sequencing. Of these amoeba positive samples, three showed no bands on agarose gel whilst 15 were overgrown with fungi and were discarded. Therefore, 43/62 (69.4 %) of the samples were positive for amoebae and of these, 38 (88.4 %) were confirmed as Vermamoeba vermiformis, four (9.2 %) as Acanthamoeba species and one (2.3 %) as Naegleria gruberi specie (GenBank accession numbers and sampling areas shown in Supplementary 1).
Phylogenetic analysis of Acanthamoeba species
Neighbor-joining (NJ) analysis was performed using the genus specific primer set JDP-1 and JDP-2 to show relationships between the Acanthamoeba positive isolates in hospital A and hospital B and reference strains from NCBI GenBank for the genotypes T1–T20. The seven Acanthamoeba isolates (GenBank accession numbers: KT385816, KT385819, KT385817, KT385814, KT385852, KT385836, KT385864) closely resembled the T3 genotype, while one isolate (GenBank accession number: KT385867) closely resembled the genotype T20 (Fig. 2).
Fig. 2.
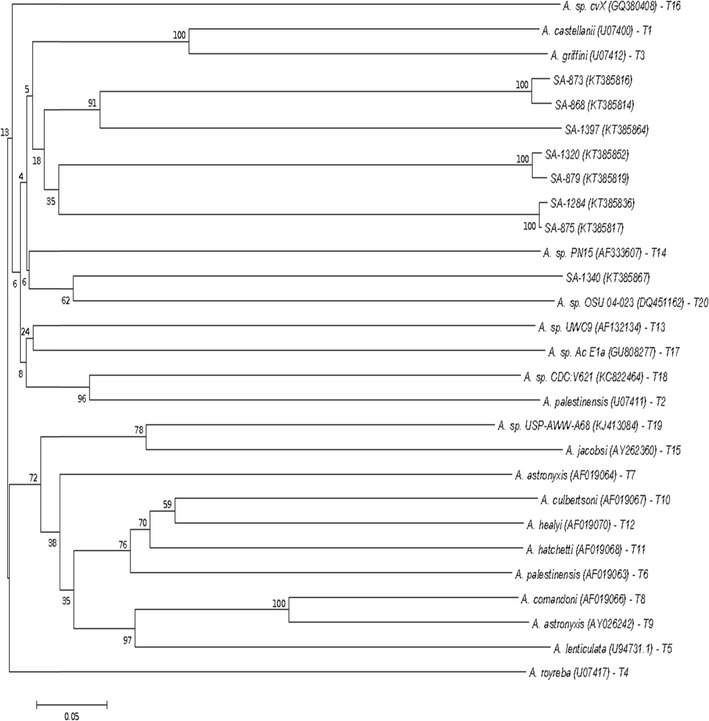
Phylogenetic relationships of Acanthamoeba species obtained from hospitals A and B and reference strains for Acanthamoeba subtype T1–T20 inferred by NJ analysis based 18S rDNA gene sequences. GenBank accession numbers of isolates and reference strains are indicated in parenthesis
Detection of amoebae-associated bacteria
Bacterial diversity in hospital A
A total of 14 isolates, composed of six bacterial species were identified using the VITEK®2 analysis from the amoebae positive samples (Table 3). The highest diversity was observed in the neonatal ward where the species Serratia marcescens, Stenotrophomonas maltophilia, Pseudomonas luteola, Rhizobium radiobacter and Archromobacter denitrificans were detected (Table 3). The most commonly detected species were Serratia marcescens (n = 5) and Stenotrophomonas maltophilia (n = 4).
Table 3.
Distribution of bacterial species at sampling areas in hospital A
| Bacterial species | Inlet (n = 4) | NW (n = 8) | NICU (n = 5) | Diarrhoea ward (n = 3) | Total |
|---|---|---|---|---|---|
| Serratia marcescens | 1 | 2 | 1 | 1 | 5 |
| Stenotrophomonas maltophilia | 0 | 3 | 1 | 0 | 4 |
| Pseudomonas luteola | 0 | 1 | 0 | 0 | 1 |
| Rhizobium radiobacter | 0 | 1 | 0 | 0 | 1 |
| Achromobacter denitrificans | 0 | 1 | 0 | 0 | 1 |
| Sphingomonas paucimobilis | 0 | 0 | 0 | 2 | 2 |
| Total | 1 | 7 | 2 | 3 | 14 |
NW neonatal ward, NICU neonatal intensive care unit
Bacterial diversity in hospital B
A total of 95 isolates composed of 21 bacterial species were detected using the VITEK®2 analysis from amoebae-positive samples of hospital B (Table 4). These bacteria were isolated from all sampling areas except the renal unit. The genera Pseudomonas and Staphylococcus were most abundant with at least two species detected at different sampling areas (Table 4). The most commonly detected species were S. marcescens (n = 25), S. maltophilia (n = 23), Delftia acidovorans (n = 9), S. paucimobilis (n = 7) and Comamonas testosteroni (n = 6).
Table 4.
Distribution of bacterial species at sampling areas in Hospital B.
| Bacterial species | CTICU (n = 3) | NICU (n = 8) | CICU (n = 4) | TICU (n = 14) | Milk room (n = 3) | EU (n = 5) | CSSD/TSSD (n = 6) | Theatre (n = 14) | Total |
|---|---|---|---|---|---|---|---|---|---|
| Sphingomonas paucimobilis | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 3 | 7 |
| Serratia marcescens | 0 | 1 | 0 | 4 | 2 | 3 | 2 | 13 | 25 |
| Stenotrophomonas maltophilia | 0 | 3 | 2 | 5 | 1 | 1 | 2 | 9 | 23 |
| Pseudomonas luteola | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| Pseudomonas fluorescens | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| Escherichia coli | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Achromobacter xylosoxiadans | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Staphylococcus hominis ssp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Staphylococcus epidermidis | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Enterococcus faecium | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Staphylococcus hominis ssp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Aerococcus viridans | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Dermacoccus nishinomiyaensis | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 4 |
| Acinetobacter lwoffii | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Aeromonas salmonicida | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Comamonas testosteroni | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 6 |
| Defiltia acidovorans | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 6 | 9 |
| Rhizobium radiobacter | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 4 |
| Kocuria varians | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| Kocuria kristinae | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Ochrobactrum anthropi | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Total | 1 | 7 | 6 | 28 | 3 | 6 | 5 | 40 | 95 |
CTICU cardio-thoracic ICU, NICU neurosurgical ICU, CICU cardiac ICU, TICU trauma ICU, EU endoscopy unit, CSSD central sterilization service unit, TSSD theatre sterilization service unit
Discussion
Physico-chemical characteristics
Generally, all the measured physico-chemical parameters were within the limits prescribed by the South African National Standard for Drinking Water Systems (SANS 241:2015). According to the standards, the accepted pH ranges are 6.0 to 9.0 and residual chlorine ≤ 5 mg/L. These standards, however, do not cover water used in haemodialysis units to treat renal patients. The Advancement of Medical Instrumentation (AAMI) used by the South African Renal Society (SARS) recommends residual chlorine of ≤ 0.50 mg/L for renal water, which is within the limits of the reverse osmosis (RO) water collected. The AAMI standards do not state the ideal pH range for renal dialysis water. Reverse osmosis (RO) membranes generally have a wide pH tolerance (2–11); however, an optimum range of 5.5–8.5 is recommended for maximum performance. The lower pH of 3.8 observed at hospital B might have been caused by acid feeds used to control membrane fouling during reverse osmosis. The resulting acidic water may leach copper from the distribution system or the dialysate which may cause side effects such as nausea, chills, headaches or even liver damage to the patient (Corbett et al. 2014).
Isolation of free-living amoebae
FLA were isolated from all the sampling areas except the milk room of hospital A and the municipal water inlet of hospital B. In total, 43.3 % (77/178) contained FLA: 50.0 % (15/30) from hospital A and 41.9 % (62/148) (35.2 %) from hospital B. The prevalence of FLA in hospital water distribution systems has been reported from previous studies in Europe, South America, the Middle East and Africa. However, there has been a lack of information regarding the occurrence of FLA from South African hospitals. The prevalence of FLA in 43.3 % (77/178) of the samples in the current study is similar with a study in Egypt by Hassan et al. (2012) which isolated amoebae in 42.9 % (30/70). Other studies in the USA and India have identified amoebae in 14.8 % (13/88) and in 14.0 % (14/100) of hospital samples, respectively (Ovrutsky et al. 2013; Khurana et al. 2015). Studies from Germany and France reported amoebae prevalence of 51.8 % (29/56) and 68.9 % (73/106) respectively (Rohr et al. 1998; Lasheras et al. 2006). The variability in the prevalence of FLA in water distribution systems can be explained by differences in geographical areas and sample source of the previous studies with the current study (Corsaro et al. 2009). In addition, there is no consensus on sampling and processing methods for isolation of amoebae from environmental samples to date, which further affects prevalence rates among studies.
The prevalence of FLA was higher in biofilm (55.4 %) than in water (32.6 %) in total samples (P > 0.05). Between the two hospitals, FLA prevalence of 38.8 % in water and 66.6 % in biofilm (P = 0.14) samples was observed for hospital A and 41.9 % water and 53.5 % in biofilm samples (P = 0.25) for hospital B. Our study is in agreement with a small study of 88 samples by Ovrutsky et al. (2013) in which FLA was observed in 15.4 % biofilm samples compared to the 13.0 % of water samples. Biofilm formation within a water distribution system can protect opportunistic pathogens, including FLA, from disinfection resulting in FLA proliferation (Barbeau and Buhler 2001; Pickup et al. 2007; Van der Wielen and Van der Kooij 2013). This might explain the high prevalence of FLA in biofilm samples compared to water samples.
Biodiversity of FLA
The genera belonging to Acanthamoeba and Vermamoeba (formerly Hartmanella) were detected in the two hospitals, with the non-pathogenic N. gruberi (GenBank accession no. KT385863) being detected only in hospital B. A study done by Doust et al. (2008) in Iran with hospital samples also observed Acanthamoeba, Vermamoeba and Naegleria species to be more prevalent. Other studies have, however, reported higher diversities of amoebae in hospital environments in comparison to the present study. A study of six hospitals in Germany with 56 hot water taps and 49 biofilm samples detected the genera Acanthamoeba, Vermamoeba, Naegleria, Vanella and Vahlkampfia (Rohr et al. 1998). Another study in a new hospital building, also in Germany reported by Michel et al. (1995), isolated Acanthamoeba, Vermamoeba, Naegleria, Echinamoeba species. Differences in diversity can also be explained by factors that include, differences in geographical areas, climate and sample type (water, biofilm or dust). Differences in methods (culture or molecular) used by study groups to identify FLA is also an important factor why different diversities are reported.
Vermamoeba vermiformis
All samples considered presumptively positive for Vermamoeba spp. after microscopic examination were confirmed by using PCR and DNA sequencing. Vermamoeba vermiformis was the most abundant amoebae species in the two hospital water systems, with a total of 48 isolates. Our findings are in agreement with a recent study in France by Pagnier et al. (2015), where 9 V. vermiformis isolates were detected from 14 amoebae species isolated in hospital water. Similar studies in Turkey and Switzerland also reported higher V. vermiformis prevalence of 72.7 and 86.7 %, respectively (Coşkun et al. 2013; Thomas et al. 2006). The genus Vermamoeba as a causative agent has been debatable, however, its isolation from human tissues suggest a role in infection (Kennedy et al. 1995; Abedkhojasteh et al. 2013; Cabello-Vílchez et al. 2014).
Acanthamoeba species
In this study, eight Acanthamoeba isolates were identified by 18S rRNA sequencing and phylogenetic analysis using the Acanthamoeba-specific primers, JDP1 and JDP2. In contrast, previous studies have reported relatively higher prevalence of Acanthamoeba spp. A study by Bagheri et al. (2010) reported the presence of Acanthamoeba spp. in 45 of the 94 (48 %) samples collected in different wards of hospitals in Iran. Two separate studies by the same research group in hospital environments also reported the presence of Acanthamoeba species in 47 of the 135 (34 %) samples collected (Carlesso et al. 2007) and 31 of the 135 (23 %) samples (Carlesso et al. 2010). Studies in Tunisia and Egypt have also shown a high detection rate of 32.6–42.9 % of Acanthamoeba species in haemodialysis and dental waters (Dendana et al. 2008; Trabelsi et al. 2012; Hassan et al. 2012). Generally, Acanthamoeba has been the most frequently isolated genera in tap water as they are easily identified morphologically compared to other FLA (Thomas et al. 2006; Coşkun et al. 2013). The low prevalence of the genus Acanthamoeba in our study relative to Vermamoeba can be attributed to the use of FLA common primers which might have been more effective in amplification of Vermamoeba rather that Acanthamoeba (Muchesa et al. 2015).
Several previous studies have shown that the T4 genotype to be connected with most Acanthamoeba infections and to be the most abundant in the hospital environment (Carlesso et al. 2010; Lasjerdi et al. 2011; Rahdar et al. 2012; Risler et al. 2013). However, in our study, phylogenetic analysis identified Acanthamoeba isolates in hospital A (neonatal ward, neonatal ICU and paediatric diarrhoea ward) and hospital B (trauma ICU, endoscopy unit and theatre) that closely resembled the T3 genotype. This was relationship was supported by a bootstrap value of 18 %. The low support values may be because of more sequences to the data set analyzed, that is, the better your data, the lower the support values (Wheeler and Pickett 2008). This means our bootstrap value may still be closer to a realistic one than other with higher figures, but few sequences. The other isolate from neurosurgical ICU of hospital B closely resembled the T20 genotype. The T3 genotype which includes A. griffin, A. pearcei and A. hatchetti has been associated with Acanthamoeba keratitis (Gonzalez-Robles et al. 2001; Visvesvara 2013). A recent study in Iran also isolated the T3 genotype in tap water and swab samples of a high dependency unit that caters for haemato-oncology patients, showing its association with hospital water systems (Khurana et al. 2015). The T20 genotype, previously miss-assigned to genotypes T16 or T4, consists of strains responsible for keratitis, granulomatous amoebic encephalitis and respiratory infections in humans (Visvesvara 2013; Fuerst et al. 2015).
Overall, the presence of the genera Vermamoeba and Acanthamoeba in hospital areas that include ICU, endoscopy unit, theatre, neonatal ward, milk kitchen, paediatric diarrhoea ward, municipal inlet and the equipment sterilization units in our study may present a potential risk factor to a large population of immunocompromised individuals and medical personnel. These individuals might be exposed to these opportunistic FLA through ingestion, inhalation or aspiration of aerosols or contact (Anaissie et al. 2002; Trabelsi et al. 2012). In addition, the residual chlorine range of 0.01–0.35 mg/L in water samples analyzed in this study is not sufficient to eliminate FLA as they have been shown to survive exposure to concentrations as high as 100 mg/L for 10 min (Storey et al. 2004). Therefore, the persistent nature of amoebal cysts means that these populations are frequently exposed to FLA.
Detection of amoebae-associated bacteria
This study investigated the biodiversity of amoebae associated bacteria in the water networks hospitals with no recent history of epidemics involving such microorganisms. Rowbotham (1980) conducted the first study to prove the association between bacteria that is Legionella and the FLA, Acanthamoeba. Numerous other studies have also shown that amoebae in man-made water systems such as drinking water, tap water, swimming pools and cooling towers can be infected by many other relevant bacteria which could use amoebae as a platform for multiplication (Greub and Raoul 2004; Molmeret et al. 2005; Thomas et al. 2006; Pagnier et al. 2008). This study is the first in South Africa where any association between free-living amoebae and bacteria has been reported in hospital water systems. In this study, the most prevalent bacterial species detected in hospital A were Serratia marcescens and S. maltophilia; hospital B: S. marcescens, S maltophilia, D. acidovorans, S. paucimobilis and C. testosteroni. All the above detected bacteria have been reported as part of the complex microbial ecology in hospital water systems. They have been implicated in hospital acquired infections in healthcare settings, with patients being exposed through drinking, skin contact and inhalation of aerosols (Thomas et al. 2006; Thomas and Ashbolt 2011; Cateau et al. 2014). These bacteria are associated with systematic blood, respiratory tract, the urinary tract, ocular and soft tissues infections (Özdemir et al. 2011; Orsini et al. 2014; Perez et al. 2014; Bilgin et al. 2015).
The amoebae, V. vermiformis, were also isolated in all samples in which S. marcescens, S. maltophilia and S. paucimobilis, D. acidovorans and C. testosteroni were detected. Our study has similarities with a study by Laganà et al. (2014), on the occurrence of FLA and Gram-negative bacteria in an Italian hospital water system, where Vermamoeba spp. coexisted with S. paucimobili, P. fluorescens and Pseudomonas aeruginosa, Acinetobacter lwoffi, S. maltophilia and C. testosteroni. The endocytobiontic relationship in all bacteria detected in this study and amoebae has been described by several studies and reviews where bacteria have been demonstrated to survive or proliferate before being released into the environment by amoebae lysis or in amoebae vesicles (Pagnier et al. 2008; Saisongkorh et al. 2010; Thomas et al. 2010; Maschio et al. 2015). Host-endozytobiont relationships in FLA allow FLA (mainly in cyst form) to form a physical barrier using the resistant cellulose cell walls, offering endozytobionts (bacteria) protection against adverse conditions such as extremes of temperature, UV light ozone, chlorine dioxide, monochloramine and copper-silver (Greub and Raoul 2004; Thomas et al. 2004; Bagheri et al. 2010). This can allow bacterial regrowth in the water system when environmental conditions are favourable (Coulon et al. 2010; Dupuy et al. 2014). In addition, low disinfectant concentrations in distribution networks have limited activity on FLA and also allow the formation of biofilms which enhances amoebae and bacteria persistence (Barbeau and Buhler 2001; Thomas et al. 2004). Studies have also shown that passage through amoebae is likely to increase the virulence and antibiotic resistance of bacteria substantially, enabling them to prepare for subsequent survival in macrophages (Miltner and Bermudez 2000; Schmitz-Esser et al. 2008). Furthermore, upregulation of virulent genes and horizontal transfer of genes can occur between the intracellular bacteria and their amoebae resulting in bacteria which are more resistant to human macrophages (Siddiqui and Khan 2012; Gomez-Valero and Buchrieser 2013).
Conclusions
Free-living amoebae were isolated from 77 (43.3 %) of the hospital samples, with 31 (32.6 %) being water and 46 (55.4 %) (P > 0.05) biofilm after amoebal enrichment. Using microscopy, PCR and 18S rDNA sequencing, Acanthamoeba spp. (T3 and T20 genotypes), V. vermiformis and Naegleria gruberi were identified. From the amoebae positive samples, S. marcescens, S. maltophilia, D. acidovorans, S. paucimobilis and C. testosteroni were the most commonly detected bacterial species. Therefore, the co-occurrence of these bacterial species with FLA in our study suggests that these interactions may be important for the transmission, survival and multiplication of bacteria in water systems where immunocompromised patients are constantly exposed, posing an important public health concern.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 22 kb)
Acknowledgments
We thank the National Institute for Occupational Health and the University of Johannesburg for providing facilities for this project and the National Research Foundation for providing a bursary. We also acknowledge the Water Research Commission for funding the project.
Compliance with ethical standards
The manuscript does not contain clinical studies or patient data.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abedkhojasteh H, Niyyati M, Rahimi F, Heidari M, Farnia S, Rezaeianr M. First report of Hartmannella keratitis in a cosmetic soft contact lens wearer in Iran. Iran J Parasitol. 2013;8:481–485. [PMC free article] [PubMed] [Google Scholar]
- Anaissie EJ, Penzak SR, Dignani C. The hospital water supply or a source of nosocomial infection: a plea for action. Arch Intern Med. 2002;162:1483–1492. doi: 10.1001/archinte.162.13.1483. [DOI] [PubMed] [Google Scholar]
- Bagheri HR, Shafiei R, Shafiei F, Sajjadi SA. Isolation of Acanthamoeba spp. from drinking waters in several hospitals of Iran. Iran J Parasitol. 2010;5:19–25. [PMC free article] [PubMed] [Google Scholar]
- Barbeau J, Buhler T. Biofilms augment the number of free-living amoebae in dental unit waterlines. Res Microbiol. 2001;152:753–760. doi: 10.1016/S0923-2508(01)01256-6. [DOI] [PubMed] [Google Scholar]
- Bilgin H, Sarmis A, Tigen E, Soyletir G, Mulazimoglu L. Delftia acidovorans: a rare pathogen in immunocompetent and immunocompromised patients. Can J Infect Dis Med Microbiol. 2015;26:277–279. doi: 10.1155/2015/973284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Vílchez AM, Mena R, Zuñiga J, Cermeño P, Martín-Navarro CM, Gonzále AC, López-Arencibia A, Reyes-Batlle M, Piñero JE, Valladares B, Lorenzo-Morales J. Endosymbiotic Mycobacterium chelonae in a Vermamoeba vermiformis strain isolated from the nasal mucosa of an HIV patient in Lima, Peru. Exp Parasitol. 2014;145:127–130. doi: 10.1016/j.exppara.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Carlesso AM, Simonetti AB, Artuso GL, Rott MB. Isolation and identification of potentially pathogenic free-living amoebae in samples from environments in a public hospital in the city of Porto Alegre, Rio Grande do Sul. Revista da Sociedade Brasileira de Medicina Tropical. 2007;40:316–320. doi: 10.1590/S0037-86822007000300013. [DOI] [PubMed] [Google Scholar]
- Carlesso A, Artuso G, Caumo K, Rott M. Potentially pathogenic Acanthamoeba isolated from a hospital in Brazil. Curr Microbiol. 2010;60:185–190. doi: 10.1007/s00284-009-9523-7. [DOI] [PubMed] [Google Scholar]
- Cateau E, Delafont V, Hechard Y, Rodier MH. Free-living amoebae: what part do they play in healthcare-associated infections? J Hos Infect. 2014;87:131–140. doi: 10.1016/j.jhin.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Corbett RW, Prout V, Haynes D, Edwards C, Frankel AH. Problems associated with hemodialysis and travel. J Trav Med. 2014;21:255–259. doi: 10.1111/jtm.12121. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Feroldi V, Saucedo G, Ribas F, Loret JF, Greub G. Novel Chlamydiales strains isolated from a water treatment plant. Environ Microbiol. 2009;11:188–200. doi: 10.1111/j.1462-2920.2008.01752.x. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Walochnik J, Kohsler M, Rott MB. Acanthamoeba misidentification and multiple labels: redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli. sp. nov. (Genotype T19) Parasitol Res. 2015;114:2481–2490. doi: 10.1007/s00436-015-4445-8. [DOI] [PubMed] [Google Scholar]
- Coşkun KA, Ozcelik S, Tutar L, Eladi N, Tutar Y. Isolation and identification of free-living amoebae from tap water in Sivas, Turkey. Biomed Res Int. 2013;2013:675145. doi: 10.1155/2013/675145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon C, Collignon A, McDonnell G, Thomas V. Resistance of Acanthamoeba cysts to disinfection treatments used in health care settings. J Clin Microbiol. 2010;48:2689–2697. doi: 10.1128/JCM.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendana F, Sellami H, Jarraya F, Sellami A, Makni F, Cheikhrouhou F, Hachicha J, Ayadi A. Free-living amoebae (FLA): detection, morphological and molecular identification of Acanthamoeba genus in the hydraulic system of an haemodialysis unit in Tunisia. Parasit. 2008;15:137–142. doi: 10.1051/parasite/2008152137. [DOI] [PubMed] [Google Scholar]
- Doust RH, Mobarez MA, Bagheri H, Khoramabadi N. Interaction of Legionellae and free-living amoebae within hospital water supplies. Res J Parasitol. 2008;3:104–113. doi: 10.3923/jp.2008.104.113. [DOI] [Google Scholar]
- Dupuy M, Berne F, Herbelin P, Binet M, Berthelot N, Rodier MH, Soreau S, Héchard Y. Sensitivity of free-living amoeba trophozoites and cysts to water disinfectants. Int J Hyg Enviro Heal. 2014;217:335–339. doi: 10.1016/j.ijheh.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Fuerst PA, Booton GC, Crary M. Phylogenetic analysis and the evolution of the 18S rRNA gene typing system of Acanthamoeba. J Eukayr Microbiol. 2015;62:69–84. doi: 10.1111/jeu.12186. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Buchrieser C (2013) Genome dynamics in Legionella: the basis of versatility and adaptation to intracellular replication. Cold Spring Harb Perspect Med 3. doi:10.1101/cshperspect.a009993 [DOI] [PMC free article] [PubMed]
- Gonzalez-Robles A, Flores-Langarica A, Omana-Molina M, Shibayama M. Acanthamoeba castellanii: ultrastructure of trophozoites using fast free fixation. J Electr Micro. 2001;50:423–427. doi: 10.1093/jmicro/50.5.423. [DOI] [PubMed] [Google Scholar]
- Greub G, Raoul D. Microorganisms resistant to free living amoebae. Clin Microbiol Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A, Farouk H, Hassanein F, Abdul-Ghani R, Abdelhady AH. Acanthamoeba contamination of hemodialysis and dental units in Alexandria, Egypt: a neglected potential source of infection. J Infect Pub Heal. 2012;5:304–310. doi: 10.1016/j.jiph.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Khurana S, Biswal M, Kaur H, Malhotra P, Arora P, Megha K, Taneja N, Sehgal R. Free living amoebae in water sources of critical units in a tertiary care hospital in India. Indian J Med Microbiol. 2015;33:343–348. doi: 10.4103/0255-0857.158543. [DOI] [PubMed] [Google Scholar]
- Laganà P, Caruso G, Piccione D, Gioffrè ME, Pino R, Delia S. Legionella spp., amoebae and not-fermenting Gram negative bacteria in an Italian university hospital water system. Ann Agric Environ Med. 2014;21:489–493. doi: 10.5604/12321966.1120623. [DOI] [PubMed] [Google Scholar]
- Lasheras A, Boulestreau H, Rogues AM, Ohayon-Courtes C, Labadie JC, Gachie JP. Influence of amoebae and physical and chemical characteristics of water on presence and proliferation of Legionella species in hospital water systems. Am J Infect Control. 2006;34:520–525. doi: 10.1016/j.ajic.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lasjerdi Z, Niyyati M, Haghighi A, Shahabi S, Biderouni FT, Taghipour N, Eftekhar M, Nazemalhosseini Mojarad E. Potentially pathogenic free-living amoebae isolated from hospital wards with immunodeficient patients in Tehran. Iranian J Parasitol. 2011;109:575–580. doi: 10.1007/s00436-011-2288-5. [DOI] [PubMed] [Google Scholar]
- Lone R, Syed K, AbuduL R, Sajjad Sheikh A, Shah F (2009) Unusual case of methicillin resistant Staphylococcus aureus and Acanthamoeba keratitis in a non-contact lens wearer from Kashmir, India. BMJ Case Rep 2009 [DOI] [PMC free article] [PubMed]
- Maschio VJ, Corção G, Rott MB. Identification of Pseudomonas spp. as amoeba-resistant microorganisms in isolates of Acanthamoeba. Revista Do Instituto De Medicina Tropical De Sao Paulo. 2015;57:81–83. doi: 10.1590/S0036-46652015000100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel R, Burghard H, Bergmann H. Acanthamoeba isolated from a highly contaminated drinking water system of a hospital exhibited natural infections with Pseudomonas aeruginosa. Zentralblatt Fur Hygiene Und Umweltmedizin. 1995;196:532–544. [PubMed] [Google Scholar]
- Miltner EC, Bermudez LE. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob Agents Chemother. 2000;44:1990–1994. doi: 10.1128/AAC.44.7.1990-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–22. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchesa P, Leifels M, Jurzik L, Barnard TG, Bartie C. Free-living amoebae isolated from a hospital water system in South Africa: a potential source of nosocomial and occupational infection. Water Sci Technol Water Supply. 2015;16:70–78. doi: 10.2166/ws.2015.106. [DOI] [Google Scholar]
- Orsini J, Tam E, Hauser N, Rajayer S (2014) Polymicrobial Bacteremia Involving Comamonas testosterone. Case Rep Med 2014 (2014), Article ID 578127, 3p [DOI] [PMC free article] [PubMed]
- Ovrutsky AR, Chan ED, Kartalija M, Bai X, Jackson M, Gibbs S, Falkinham JO, Iseman MD, Reynolds PR, McDonnell G. Co-occurrence of free-living amoebae and nontuberculous Mycobacteria in hospital water networks, and preferential growth of Mycobacterium avium in Acanthamoeba lenticulata. Appl Environ Microbiol. 2013;79:3185–3192. doi: 10.1128/AEM.03823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdemir M, Pekcan S, Demircili ME, Taşbent FE, Feyzioğlu B, Pirinç S, Baykan M. A rare cause of bacteremia in a pediatric patient with Down syndrome: Sphingomonas paucimobilis. Int J Med Sci. 2011;8:537–539. doi: 10.7150/ijms.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozerol IH, Bayraktar M, Cizmeci Z, Durmaz R, Akbas E, Yildirim Z, Yologlu S. Legionnaire’s disease: a nosocomial outbreak in Turkey. J Hosp Infect. 2006;62:50–57. doi: 10.1016/j.jhin.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Pagnier I, Raoult D, La Scola B. Isolation and identification of amoeba-resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ Microbiol. 2008;10:1135–1144. doi: 10.1111/j.1462-2920.2007.01530.x. [DOI] [PubMed] [Google Scholar]
- Pagnier I, Yutin N, Croce O, Makarova KS, Wolf YI, Benamar S, Raoult D, Koonin EV, La Scola B. Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biol Dir. 2015;10:13. doi: 10.1186/s13062-015-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez PN, Ramirez M, Fernandez JA, De Guevara LL. A patient presenting with cholangitis due to Stenotrophomonas maltophilia and Pseudomonas aeruginosa successfully treated with intrabiliary colistin. Curr Infect Dis Rep. 2014;6:5147. doi: 10.4081/idr.2014.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup ZL, Pickup R, Parry JD. Growth of Acanthamoeba castellanii and Hartmannella vermiformis on live, heat-killed and DTAF-stained bacterial prey. FEMS Microbiol Ecol. 2007;61:264–272. doi: 10.1111/j.1574-6941.2007.00346.x. [DOI] [PubMed] [Google Scholar]
- Rahdar M, Niyyati M, Salehi M, Feghhi M, Makvandi M, Pourmehdi M, Farnia S. Isolation and genotyping of Acanthamoeba strains from environmental sources in Ahvaz City, Khuzestan Province, Southern Iran. Iranian J Parasitol. 2012;7:22–26. [PMC free article] [PubMed] [Google Scholar]
- Risler A, Coupat-Goutaland B, Pélandakis M. Genotyping and phylogenetic analysis of Acanthamoeba isolates associated with keratitis. Parasitol Res. 2013;112:3807–3816. doi: 10.1007/s00436-013-3572-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Zaragoza S. Ecology of free-living amoebae. Crit Rev in Microbiol. 1994;20:225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- Rohr U, Weber S, Michel R, Selenka F, Wilhelm M. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl EnvironMicrobiol. 1998;64:1822–1824. doi: 10.1128/aem.64.5.1822-1824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisongkorh W, Robert C, La Scola B, Raoult D, Rolain JM. Evidence of transfer by conjugation of type IV secretion system genes between Bartonella Species and Rhizobium radiobacter in Amoeba. PLoS ONE. 2010;5:e12666. doi: 10.1371/journal.pone.0012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid P. Relevance of free-living amoebae as hosts for phylogenetically diverse microorganisms. Parasitol Res. 2014;113:2407–2414. doi: 10.1007/s00436-014-3932-7. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, Toenshoff ER, Haider S, Heinz E, Hoenninger VM, Wagner M, Horn M. Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl Environ Microbiol. 2008;74:5822–5831. doi: 10.1128/AEM.01093-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South African National Standard (SANAS) 241 (2015) Drinking water. Part1: Microbiological, physical aesthetic and chemical determinants.
- Stojek NM, Szymańska J, Dutkiewicz J. Gram-negative bacteria in water distribution systems of hospitals. Ann Agric Environ Med. 2008;15:135–142. [PubMed] [Google Scholar]
- Storey MV, Winiecka-KrusnellI J, Ashbolt NJ, Stenstorm TA. The efficacy of heat and chlorine treatment against thermotolerant Acanthamoebae and Legionellae. Scand J Infect Dis. 2004;36:656–662. doi: 10.1080/00365540410020785. [DOI] [PubMed] [Google Scholar]
- Taylor GT. The role of pelagic heterotrophic protozoa in nutrient cycling: a review. Ann Inst Oceanogr. 1982;58:227–241. [Google Scholar]
- Thomas JM, Ashbolt NJ. Do free-living amoebae in treated drinking water systems present an emerging health risk. Environ Sci Technol. 2011;45:860–869. doi: 10.1021/es102876y. [DOI] [PubMed] [Google Scholar]
- Thomas V, Bouchez T, Nicholas V, Robert S, Loret JF, Levi Y. Amoebae in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence. Appl Environ Microbiol. 2004;97:950–963. doi: 10.1111/j.1365-2672.2004.02391.x. [DOI] [PubMed] [Google Scholar]
- Thomas V, Herrera-Rimann K, Blannci DS, Greub G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl Environ Microbiol. 2006;72:2428–2438. doi: 10.1128/AEM.72.4.2428-2438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi H, Dendana F, Sellami A, Sellami H, Cheikhrouhou F, Neji S, Makni F, Ayadi A. Pathogenic free-living amoebae: epidemiology and clinical review. Pathologie Biologie. 2012;60:399–405. doi: 10.1016/j.patbio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Van der Wielen PWJJ, van der Kooij D. Non-tuberculosis Mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in the Netherlands. Appl Environ Microbiol. 2013;79:825–834. doi: 10.1128/AEM.02748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvesvara GS. Infections with free-living amoebae. Hand Clinical Neurol. 2013;114:153–168. doi: 10.1016/B978-0-444-53490-3.00010-8. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Moura H, Shuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Wheeler WC, Pickett KM. Topology-Bayes versus Clade-Bayes in phylogenetic analysis. Mol Biol Evol. 2008;25:447–453. doi: 10.1093/molbev/msm274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 22 kb)


