Abstract
Biosimilars of granulocyte colony-stimulating factor (G-CSF) have been routinely introduced into clinical practice. However, not functional genomics characterization has been performed yet in comparison with the innovator G-CSF. This study aimed to evaluate the transcriptomic changes in an in vitro model of umbilical cord blood cells (UBC) exposed to G-CSF for the identification of their modulated pathways. Umbilical cord blood cells–derived mononuclear cells (MNCs) were treated with biosimilar and innovator G-CSF for further gene expression profiling analysis using a microarray-based platform. Comparative analysis of biosimilar and innovator G-CSF gene expression signatures allowed us to identify the most commonly modulated pathways by both drugs. In brief, we observed predominantly upmodulation of transcripts related to PI3K-Akt, NF-kappaB, and tumor necrosis factor (TNF) signaling pathways as well as transcripts related to negative regulation of apoptotic process among others. In addition, hematopoietic colony-forming cell assays corroborate the G-CSF phenotypic effects over UBC-derived MNCs. In conclusion, our study suggests that G-CSF impacts UBC-derived cells through the modulation of several signaling pathways associated with cell survival, migration, and proliferation. The concordance observed between biosimilar and innovator G-CSF emphasizes their similarity in regards to their specificity and biological responses.
Keywords: G-CSF, innovator, biosimilar, umbilical cord blood, transcriptomics
Introduction
Granulocyte colony-stimulating factor (G-CSF) is a growth factor that modulates hematopoiesis and the immune system. It is widely used to induce myelosuppression, proliferation, and differentiation of hematopoietic cells and to activate the neutrophil functions in clinical practice as well as in radiotherapy and chemotherapy. Furthermore, it is also administered to healthy donors to mobilize stem cells from the bone marrow to the peripheral blood. Granulocyte colony-stimulating factor exerts its biological effects through binding to its receptor CSF3R (CD114), which is expressed in hematopoietic stem cells (HSC), neutrophil precursors, mature neutrophils, and myeloid cell lines. It has been described that G-CSF affects the immune system by modifying the reactivity of T cells and the cellular function of antigen presentation. Several studies have shown that G-CSF stimulates the inflammatory response while suppressing the adaptive immune system.1
Currently, the G-CSF patent that is produced by biotechnology has expired, leading to the development of similar drugs by non-innovative medicines known as biosimilars. The use of the innovator and biosimilar G-CSF has been widely extended for the neutropenia treatment produced by chemotherapy. However, the use of G-CSF in the mobilization of HSC to the peripheral blood is carried out preferentially by innovative medicine.2 Thus, new studies are needed to provide more evidence on both efficacy and safety to assess the quality of these new biosimilar G-CSFs. The confirmation of these results will impact their associated cost reduction due to the questions regarding non-equality and non-bioequivalence, which could influence the sustainability of the health system.
Gene expression changes after exposure to an in vitro drug can provide a comprehensive characterization and estimation of its bioequivalence properties. The transcriptomics analysis as an approach to evaluate the toxicological profile of drugs was introduced more than 20 years ago,3 and it has been widely applied to the pharmacological relevance field.4 Furthermore, the use of these genomic approaches could be used to assess the risks and efficacy of biosimilar medicines.1
This study aimed to evaluate the transcriptomic changes in an in vitro model of umbilical cord blood cells (UBC) exposed to the biosimilar and innovator G-CSF for the identification of their modulated pathways.
Materials and Methods
Umbilical cord blood samples and derived mononuclear cells
A total of 41 UBC samples were obtained in the gynecology service of the Central Military Hospital and 13 UBC donated by the Public Bank of the Capital District of Bogotá. After the birth of the newborn, umbilical cord clamping was performed, and after disinfection with iodopovidone at the site, the umbilical cord vein was punctured. Blood was collected by gravity in a Terumo® bag. None of the mothers involved reported a history of infectious and contagious diseases (HIV, Hepatitis B, Hepatitis C, or Syphilis) or complications during the birth of the newborn. Umbilical cord blood cells units were analyzed for nucleated cell counts, and the cells were subsequently processed by density gradient separation 1077 (Ficoll-Hypaque) to obtain mononuclear cells (MNC); 20 out of 54 samples were selected because of a greater than 95% viability and a number of MNC greater than 1 × 106 cells. The selected samples were counted for CD34+ by flow cytometry (FacsCanto II; Becton Dickinson, Franklin Lakes, NJ, USA). The stem cell enumeration BD™ kit was used by the ISHAGE protocol (Becton Dickinson, Franklin Lakes, NJ, USA), and UBC subpopulations were characterized using monoclonal antibodies: CD114 PE-A, CD44 FITC-A, CD133 APC-A, CD105 PerCP-A, CD90 FITC-A, CD73 PE-A, CD19 PE-A, CD34/45, and CD38/45, following manufacturer’s protocols. Umbilical cord blood cells–derived MNCs were cryopreserved at −196°C until use.
The Institutional Ethics Committee of the Hospital Militar Central approved this study under code 2013-049, according to the World Medical Association Declaration of Helsinki. Pregnant women donated human UBC after signing informed consent.
Stimulation of UBC-derived MNCs with biosimilar and innovator G-CSFs
The optimal dose (100 or 200 ng/mL) and treatment time point (0, 8, 24, and 48 hours) of the innovator (Neupogen; Roche, Basel, Switzerland) and biosimilar (IOR®leukoCIM; Delta Labs, Bogotá, Colombia) were evaluated using the 7AAD cell viability staining assay by flow cytometry on pooled UBC-derived MNCs. Based on the resulting data, subsequent experiments were performed with 100 ng/mL at 6 hours after stimulation to capture early gene expression changes without compromise of cell viability. Transcriptomics profiling was determined with pooled UBC-derived MNCs from different individuals in duplicate tests. Identical cell concentrations were subjected or not (control) to pharmacological stimulation with the innovator or biosimilar G-CSF in 3 independent experiments.
CD114+ expression analysis by flow cytometry
Umbilical cord blood cells was seeded in 6 × 6 plates at a concentration of 1 × 105 MNCs and was stimulated with 100 ng of both the innovative and biosimilar. Filgrastim expression of the CD114 receptor was measured prior to the stimulus and 6 hours after. Antibodies against human antigens CD114 PE-A were purchased from Becton Dickinson. A total of 1 × 105 cells/mL cells were resuspended in 0.2 mL Dulbecco phosphate-buffered saline (DPBS) and incubated with fluorescein isothiocyanate (FITC-conjugated antibodies for 30 minutes at room temperature. The fluorescence intensity of the cells was evaluated by flow cytometry (FACS Canto II; Becton Dickinson, Franklin Lakes, NJ, USA), and the data were analyzed using the FACS Diva software (Becton Dickinson, Franklin Lakes, NJ, USA).
Hematopoietic colony-forming cell assays
The evaluation of stem and progenitor cells as colony-forming units (CFU) was assessed in semi-solid cultures based on methylcellulose (StemMACS HSC-FU lite with Epo, human) in Iscove medium (IMDM) and supplemented with fetal bovine serum (FBS) and different growth factors. The procedure was performed according to the supplier’s protocol. Umbilical cord blood cells–derived MNCs were adjusted to the recommended concentration and read after 15 days of culture.
RNA isolation and transcriptomics analysis
Total RNA was isolated from 12 pooled UBC-derived MNCs obtained from 20 different individuals using the Quick-RNA™ MiniPrep Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. The RNA quality and concentration were determined with Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). Two micrograms of total RNA derived from UBCs treated with G-CSF were labeled with Cy3, while untreated counterparts were labeled with Cy5 (used as control) for their further hybridization on Agilent SurePrint G3 Human GE v2 8x60K Microarray (Agilent Technologies, Palo Alto, CA, USA). The hybridization steps were carried out according to the Agilent protocol, and images were acquired using a Genepix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA). Image analysis and initial quality control were performed using Agilent Feature Extraction Software v10.2. Raw and pre-processed data have been deposited in the Gene Expression Omnibus (GEO) repository with the identifier GSE139352.
Limma Bioconductor package was employed for background adjustment, within and between arrays normalization. Eight out of 12 generated gene expression profiles were selected for their further analysis based on outlier analysis. We employed the one-class Rank Products’ test (P value < .01; fold change > 1.5) for differential gene expression analysis using the MultiExperiment Viewer software (MeV 4.9).5 The number and identity of genes commonly affected in both drugs were determined with Venn mapping.6 Functional enrichment analyses were performed with Enrichr (https://amp.pharm.mssm.edu/Enrichr/) and NetworkAnalyst (https://www.networkanalyst.ca).
Statistical analysis
Statistical analysis of flow cytometry data (CD114+ expression) and the colony-forming cell assays were performed using R software, applying Mann-Whitney test or Kruskal-Wallis test as appropriate. A P < .05 was considered statistically significant.
Results and Discussion
Recently, biosimilars of G-CSF drugs have been routinely introduced into clinical practice to mobilize CD34+ cells and accelerate engraftment after transplantation.7 Biosimilar products have the potential to help decrease the price and increase the availability of biological medicines. However, the demonstration of similarity and comparability of drugs is based on extensive structural and functional characterization, including primary sequence analysis, higher order protein structure, post-translational modifications, protein aggregation, product-related impurities, and biological activities.8 Comparisons of the different G-CSF exist only in the clinical setting that does not allow the head to head or molecular comparisons. Transcriptomics-based approaches have been proposed as a generalized framework for the assessment of biosimilar biological properties with respect to the innovator product.1,9-13
The primary goal of our study was to identify commonly deregulated pathways among biosimilar and innovator G-CSF in UBC-derived cells. To this end, gene expression data based on a microarray platform was obtained from pooled UBC-derived MNCs after 6 hours of treatment with biosimilar or innovator G-CSFs and their corresponding untreated counterparts samples used as controls. First, we performed a comprehensive evaluation of gene expression profiles associated with biosimilar and innovator G-CSF treatments, to identify the most representative differentially expressed transcripts for each drug. Second, we compared the biosimilar and innovator G-CSF gene expression signatures to identify the commonly modulated transcripts by both drugs. Third, functional enrichment analysis was performed to detect the G-CSF-modulated pathways in UBC-derived MNCs.
Characterization of UBC-derived MNCs surface markers
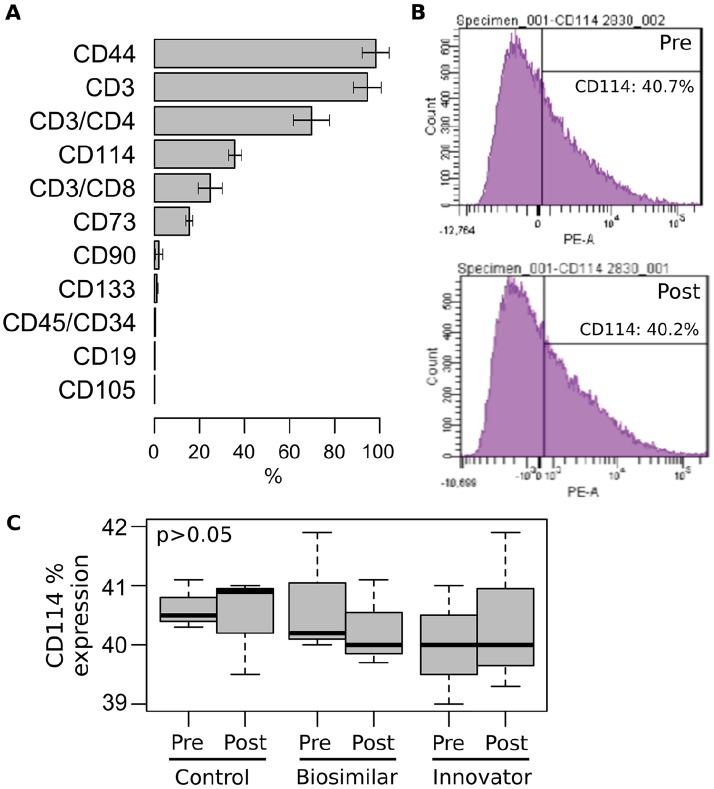
Before transcriptomics analysis, UBC-derived samples were characterized by flow cytometry analysis of 12 different surface markers (Figure 1A). The most frequently expressed markers among the UBC-derived samples were CD44 (98.2%), CD3 (94.8%), CD3/CD4 (69.8%), CD114 (35.7%), CD3/CD8 (24.7%), and CD73 (15.5%). In addition, CD114 expression analysis of pooled biological replicates in pre- and post-treatment conditions showed no statistically significant differences (P > .05; Figure 1B and C). Interestingly, the cellular heterogeneity identified in the UBC-derived MNCs populations (eg, T and B cells, MSCs, cytopathic effect [CPE]) are in agreement with previous studies.14-18 Recently, Zheng et al performed Drop-seq and scRNA-seq to dissect cellular heterogeneity in CD34+ progenitors in UBC. This study identified a “striking heterogeneity” in UBC cell subsets in the early molecular transitions toward the commitment of distinct myeloid cells.19
Figure 1.
Surface markers analysis of UBC-derived MNCs. (A) Flow cytometry analysis of 12 different surface markers. The average percentage of expression (±SD) for the UBC samples are shown. (B) Flow cytometry analysis of CD114 expression. (C) Comparative analysis of CD114 expression among UBC-derived MNCs pre- and post-stimulated with biosimilar and innovator G-CSF. Experiments were performed in triplicate and compared with Mann-Whitney test. G-CSF indicates granulocyte-colony-stimulating factor; MNC, mononuclear cells; UBC, umbilical cord blood cells.
G-CSF transcriptomics modulation on UBC-derived MNCs
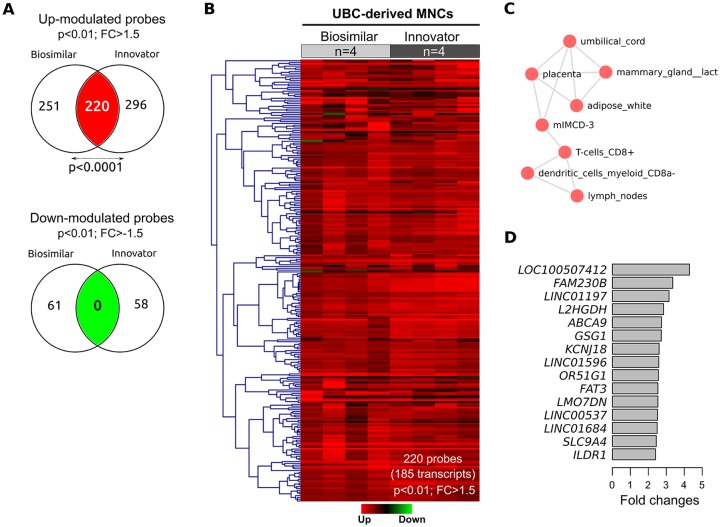
A global gene expression profiling was carried out in UBC-derived MNCs treated with either biosimilar and innovator G-CSF with respect to untreated controls. We identified 532 and 574 probes, respectively, modulated by biosimilar and innovator G-CSF (fold change ⩾ 1.5; P < .01; Figure 2A). Comparative analysis of their gene expression signatures revealed 185 transcripts upmodulated (220 probes) across biosimilar and innovator G-CSF, representing a non-random significant number (40% probes) of overlapping genes based on the normal approximation to the binomial distribution (P < .0001; Figure 2A; Supplementary data 1). These results demonstrated a significant similarity between biosimilar and innovator G-CSFs at transcriptomics level (Figure 2B) modulating the expression of UBC-related genes, among others (Figure 2C). Interestingly, the most commonly overexpressed transcripts in G-CSF treatments were several uncharacterized long intergenic non-coding RNAs (eg, LINC01197, LINC01596, LINC00537, and LINC01684; Figure 2D). In addition, comparison of gene expression on biosimilar and innovator G-CSF treatment revealed a significant number of transcripts differentially repressed by both drugs as shown in the Venn diagram of downmodulated probes (Figure 1A).
Figure 2.
Transcriptomics analyses of G-CSF biosimilar and innovator on UBC-derived MNCs. (A) Identification of transcripts commonly modulated by biosimilar and innovator G-CSF (P ⩽ .01, fold change ⩾ 1.5). (B) Heatmap of commonly upmodulated transcripts among biosimilar and innovator G-CSF. Gene expression levels are shown with a green/red color scale, red indicating upmodulation. (C) Enrichment analysis of the most representative cells according to genes commonly upmodulated by both drugs. (D) Top 15 upmodulated genes based on fold changes. G-CSF indicates granulocyte-colony stimulating factor; MNC, mononuclear cells; UBC, umbilical cord blood cells.
Functional enrichment analysis of commonly deregulated transcripts revealed specific bioprocess related to transmembrane transport of small molecules (P = .001), negative regulation of apoptotic process (P = .018), phospholipase C activating G-protein-coupled receptor (GPCR) signaling (P = .02), PI3K-Akt signaling pathway (P = .028), activation of nuclear factor kappa B (NF-kappaB; P = .03) and tumor necrosis factor (TNF) signaling pathways (P = .049) among others (Figure 3A). Furthermore, functional enrichment analysis of genes exclusively modulated in each drug revealed specific metabolic and cell-cell communication process enriched in the biosimilar treatment (P < .03; Figure 3B), while innovator G-CSF was characterized by the modulation of transforming growth factor (TGF)-beta signaling pathway (P = .001), leukocyte migration (P = .002), p53 signaling pathway (P = .004), positive regulation mitogen-activated protein (MAP) kinase activity (P = .01) and negative regulation of cell adhesión (P = .01) among others (Figure 3C). It is important to note that differences in the modulation of these specific pathways by G-CSF formulations could derive in different clinical implications and/or side effects that deserve further analysis.
Figure 3.
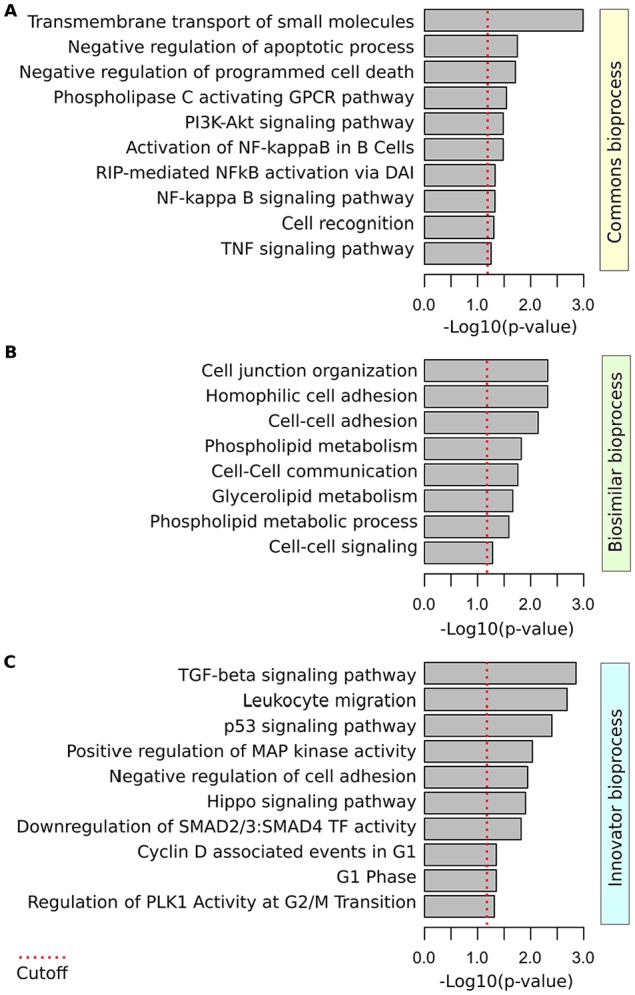
Functional enrichment analysis of G-CSF-modulated bioprocess in UBC-derived MNCs. (A) Bioprocess enriched in biosimilar and innovator G-CSF common gene expression signature. (B) Bioprocess enriched among the gene exclusively modulated by the biosimilar G-CSF. (C) Bioprocess enriched among the gene exclusively modulated by the innovator G-CSF. G-CSF indicates granulocyte-colony-stimulating factor; MNC, mononuclear cells; UBC, umbilical cord blood cells; TNF, tumor necrosis factor; TGF, transforming growth factor; GPCR, G-protein-coupled receptor; RIP, receptor-interacting serine/threonine-protein; MAP, mitogen-activated protein.
Granulocyte colony-stimulating factor (G-CSF) modulation of PI3K/Akt signaling pathways on UBC-derived cells could be protecting cells from death through the upregulation of anti-apoptotic proteins.20 Notably, both G-CSF compounds regulate a subset of genes related to negative regulation of apoptotic process (eg, BNIP3L, GCLM, KRT18, PSMD10, WNT1, CFLAR, C8orf44-SGK3), which suggests protection against apoptosis and increased cell survival. Anti-apoptotic G-CSF effects have been previously described in several studies. Philpott et al21 showed that G-CSF not only blocks apoptosis but also prolongs the survival of peripheral blood neutrophils. Schmidt-Mende et al22 have shown that G-CSF has anti-apoptotic properties through caspase activity inhibition in hematopoietic progenitor and bone marrow cells isolated from refractory anemia patients.
The involvement of PI3K/Akt as a central signaling pathway in uterine serous carcinomas (USCs) has been previously postulated by Furmento and co-workers, demonstrating its role in human trophoblast cell survival, proliferation, and migration.23,24 In this sense, our analysis identified the upmodulation of several transcripts (eg, C8orf44-SGK3, GNG13, IKBKB, LAMA5) involved with the regulation of cell migration and cytoskeletal organization through PI3K and Rho proteins signaling pathways.25-27 Ponte et al28 showed that G-CSF-stimulated migration was mediated by a mechanism that involves metalloproteinases activation. Also, G-CSF-induced morphological changes in actin cytoskeleton consistent with a migratory cell phenotype of human trophoblast Swan 71 cells via PI3K and mitogen-activated protein kinase (MAPK) activation.29
To corroborate the G-CSF phenotypic effects over cell differentiation, proliferation, and migration, a hematopoietic colony-forming cell assay was performed on an independent group of UBC-derived MNCs samples (Figure 4A). These results demonstrated a significantly increased number of colony-forming unit—erythroid and colony-forming unit—granulocyte/monocyte after stimulation with biosimilar and innovator G-CSFs compared with their controls (P < .01; Figure 4B). In addition, CFU test showed no statistically significant differences between biosimilar and innovator G-CSF over their capability for the differentiation of UBC cells toward white colonies enriched with granulocytes.
Figure 4.
Hematopoietic colony-forming cell assays. (A) Microphotographs of CFU-E, CFU-GM, CFU-Mix, and hematoxilyn-eosin staining among untreated and treated UBC-derived cells. (B) Statistical analysis of colony-forming assay was based on the average counts of five UBC-derived MNCs samples stimulated with biosimilar or innovator G-CSF and compared with the control using Kruskal-Wallis test. CFU indicates colony-forming units; G-CSF, granulocyte-colony stimulating factor; MNC, mononuclear cells; UBC, umbilical cord blood cells.
Transcriptomics characterization suggests that G-CSF mainly serves as a pro-survival factor for UBC-derived cells through the activation of PI3K/Akt and inactivation apoptotic signaling pathways. Moreover, our functional genomics analysis suggests that both G-CSF drugs could be involved with the innate immune response due to the modulation of genes related to the NF-kappa B and TNF signaling pathways (eg, CFLAR, IKBKB, PSMD10). On the other hand, Human and animal model studies have demonstrated that G-CSF stimulation alters the function of T cells and modulates the balance between Th1 and Th2 immune responses by affecting cytokine production.30
In conclusion, our study suggests that G-CSF impacts UBC-derived cells through the modulation of several signaling pathways associated with cell survival, migration, and proliferation, recapitulating previous results obtained from in vivo models. The concordance observed between biosimilar and innovator G-CSF emphasizes their similarity in regards to their specificity and biological responses despite the cellular heterogeneity of UBC-derived samples, genetic diversity of donors, and cell differentiation states. Nevertheless, further studies should be conducted in other in vitro and in vivo models to corroborate our findings and also to elucidate the similarity and differences in the clinical relevances of both G-CSF formulations.
Supplemental Material
Supplemental material, Supplementary_data_1_xyz323131a700f5f for Comparative Analysis of the Biosimilar and Innovative G-CSF Modulated Pathways on Umbilical Cord Blood–Derived Mononuclear Cells by LM Avila-Portillo, F Aristizabal, S Perdomo, A Riveros, B Ospino, JP Avila, M Butti and MC Abba in Bioinformatics and Biology Insights
Footnotes
Funding:The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by Hospital Militar Central (Bogotá, Colombia).
Declaration of Conflicting Interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Luz M. Avila-Portillo holds a position as scientific director of Stem Medicina Regenerativa/CryoHoldco in Bogotá, Colombia.
Author Contributions: LMA, FA and MCA designed research. LMA, SP, AR, BO and JPA performed research. LMA, MB and MCA analyzed the data. LMA and MCA wrote the paper.
ORCID iDs: LM Avila-Portillo  https://orcid.org/0000-0002-1734-6361
https://orcid.org/0000-0002-1734-6361
Supplemental Maerial: Supplemental material for this article is available online.
References
- 1. Welte K. G-CSF: filgrastim, lenograstim and biosimilars. Expert Opin Biol Ther. 2014;14:983-993. [DOI] [PubMed] [Google Scholar]
- 2. Kurki P, van Aerts L, Wolff-Holz E, Giezen T, Skibeli V, Weise M. Interchangeability of biosimilars: a European perspective. BioDrugs. 2017;31:83-91. doi: 10.1007/s40259-017-0210-0. [DOI] [PubMed] [Google Scholar]
- 3. Chen M, Zhang M, Borlak J, Tong W. A decade of toxicogenomic research and its contribution to toxicological science. Toxicol Sci. 2012;130:217-228. doi: 10.1093/toxsci/kfs223. [DOI] [PubMed] [Google Scholar]
- 4. Goetz AK, Singh BP, Battalora M, et al. Current and future use of genomics data in toxicology: opportunities and challenges for regulatory applications. Regul Toxicol Pharmacol. 2011;61:141-153. doi: 10.1016/j.yrtph.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 5. Saeed A, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374-378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 6. Smid M, Dorssers LC, Jenster G. Venn mapping: clustering of heterologous microarray data based on the number of co-occurring differentially expressed genes. Bioinformatics. 2003;19:2065-2071. doi: 10.1093/bioinformatics/btg282. [DOI] [PubMed] [Google Scholar]
- 7. Yoshimura H, Hotta M, Nakanishi T, et al. Evaluation of a biosimilar granulocyte colony-stimulating factor (filgrastim XM02) for peripheral blood stem cell mobilization and transplantation: a single center experience in Japan. J Blood Med. 2017;8:5-12. doi: 10.2147/JBM.S123374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsuruta LR, Lopes dos Santos M, Moro AM. Biosimilars advancements: moving on to the future. Biotechnol Prog. 2015;31:1139-1149. doi: 10.1002/btpr.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pabst T, Vellenga E, van Putten W, et al. ; and Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON); German AML Study Group (AMLSG); Swiss Collaborative Group for Clinical Cancer Research (SAKK). Favorable effect of priming with granulocyte colony-stimulating factor in remission induction of acute myeloid leukemia restricted to dose escalation of cytarabine. Blood. 2012;119:5367-5373. doi: 10.1182/blood-2011-11-389841. [DOI] [PubMed] [Google Scholar]
- 10. Taylor KM, Jagannath S, Spitzer G, et al. Recombinant human granulocyte colony-stimulating factor hastens granulocyte recovery after high-dose chemotherapy and autologous bone marrow transplantation in Hodgkin’s disease. J Clin Oncol. 1989;7:1791-1799. doi: 10.1200/JCO.1989.7.12.1791. [DOI] [PubMed] [Google Scholar]
- 11. Sheridan WP, Morstyn G, Wolf M, et al. Granulocyte colony-stimulating factor and neutrophil recovery after high-dose chemotherapy and autologous bone marrow transplantation. Lancet. 1989;2:891-895. doi: 10.1016/s0140-6736(89)91552-3. [DOI] [PubMed] [Google Scholar]
- 12. Bonilla MA, Gillio AP, Ruggeiro M, et al. Effects of recombinant human granulocyte colony-stimulating factor on neutropenia in patients with congenital agranulocytosis. N Engl J Med. 1989;320:1574-1580. doi: 10.1056/NEJM198906153202402. [DOI] [PubMed] [Google Scholar]
- 13. Karagianni N, Kranidioti K, Fikas N, et al. An integrative transcriptome analysis framework for drug efficacy and similarity reveals drug-specific signatures of anti-TNF treatment in a mouse model of inflammatory polyarthritis. PLoS Comput Biol. 2019;15:e1006933. doi: 10.1371/journal.pcbi.1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sorg RV, Andres S, Kögler G, Fischer J, Wernet P. Phenotypic and functional comparison of monocytes from cord blood and granulocyte colony-stimulating factor-mobilized apheresis products. Exp Hematol. 2001;29:1289-1294. doi: 10.1016/s0301-472x(01)00735-4. [DOI] [PubMed] [Google Scholar]
- 15. Mayani H, Lansdorp PM. Biology of human umbilical cord blood-derived hematopoietic stem/progenitor cells. Stem Cells. 1998;16:153-165. doi: 10.1002/stem.160153. [DOI] [PubMed] [Google Scholar]
- 16. Secco M, Zucconi E, Vieira NM, et al. Multipotent stem cells from umbilical cord: cord is richer than blood! Stem Cells. 2008;26:146-150. doi: 10.1634/stemcells.2007-0381. [DOI] [PubMed] [Google Scholar]
- 17. Zuba-Surma EK, Klich I, Greco N, Laughlin MJ, Ratajczak J, Ratajczak MZ. Optimization of isolation and further characterization of umbilical-cord-blood-derived very small embryonic/epiblast-like stem cells (VSELs). Eur J Haematol. 2010;84:34-46. doi: 10.1111/j.1600-0609.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 18. Domanska-Janik K, Buzanska L, Lukomska B. A novel, neural potential of non-hematopoietic human umbilical cord blood stem cells. Int J Dev Biol. 2003;52:237-248. doi: 10.1387/ijdb.072315kd. [DOI] [PubMed] [Google Scholar]
- 19. Zheng S, Papalexi E, Butler A, Stephenson W, Satija R. Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol. 2018;14:e8041. doi: 10.15252/msb.20178041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liongue C, Wright C, Russell AP, Ward AC. Granulocyte colony-stimulating factor receptor: stimulating granulopoiesis and much more. Int J Biochem Cell Biol. 2009;41:2372-2375. doi: 10.1016/j.biocel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 21. Philpott NJ, Prue RL, Marsh J, Gordon-Smith EC, Gibson FM. G-CSF-mobilized CD34 peripheral blood stem cells are significantly less apoptotic than unstimulated peripheral blood CD34 cells: role of G-CSF as survival factor. Br J Haematol. 1997;97:146-152. doi: 10.1046/j.1365-2141.1997.d01-2126.x. [DOI] [PubMed] [Google Scholar]
- 22. Schmidt-Mende J, Tehranchi R, Forsblom AM, et al. Granulocyte colony-stimulating factor inhibits fas-triggered apoptosis in bone marrow cells isolated from patients with refractory anemia with ringed sideroblasts. Leukemia. 2001;15:742-751. doi: 10.1038/sj.leu.2402110. [DOI] [PubMed] [Google Scholar]
- 23. Furmento VA, Blank VC, Madhivanan K, Aguilar RC, Marino VJ, Roguin LP. MAPKs and PI3K/akt pathways are involved in G-CSF-induced migration in a human trophoblast cell line. Placenta. 2015;4:488. doi: 10.1016/j.placenta.2015.01.445. [DOI] [Google Scholar]
- 24. New DC, Wu K, Kwok AW, Wong YH. G protein-coupled receptor-induced Akt activity in cellular proliferation and apoptosis. FEBS J. 2007;274:6025-6036. doi: 10.1111/j.1742-4658.2007.06116.x. [DOI] [PubMed] [Google Scholar]
- 25. Pinto JB. Role of Novel Nuclear Envelope Proteins Involved in Nuclear Positioning during Cell Migration [master’s thesis]. Lisbon: Universidade de Lisboa; 2010. [Google Scholar]
- 26. Yamazaki T, Yang XO, Chung Y, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181:8391-8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kariya Y, Miyazaki K. The basement membrane protein laminin-5 acts as a soluble cell motility factor. Exp Cell Res. 2004;297:508-520. doi: 10.1016/j.yexcr.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 28. Ponte AL, Ribeiro-Fleury T, Chabot V, et al. Granulocyte-colony-stimulating factor stimulation of bone marrow mesenchymal stromal cells promotes CD34+ cell migration via a matrix metalloproteinase-2-dependent mechanism. Stem Cells Dev. 2012;21:3162-3172. doi: 10.1089/scd.2012.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furmento VA1, Marino J, Blank VC, et al. Granulocyte colony-stimulating factor (G-CSF) upregulates β1 integrin and increases migration of human trophoblast Swan 71 cells via PI3K and MAPK activation. Exp Cell Res. 2016;342:125-134. doi: 10.1016/j.yexcr.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buzzeo MP, Yang J, Casella G, Reddy V. Hematopoietic stem cell mobilization with G-CSF induces innate inflammation yet suppresses adaptive immune gene expression as revealed by microarray analysis. Exp Hematol. 2007;35:1456-1465. doi: 10.1016/j.exphem.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_data_1_xyz323131a700f5f for Comparative Analysis of the Biosimilar and Innovative G-CSF Modulated Pathways on Umbilical Cord Blood–Derived Mononuclear Cells by LM Avila-Portillo, F Aristizabal, S Perdomo, A Riveros, B Ospino, JP Avila, M Butti and MC Abba in Bioinformatics and Biology Insights