Abstract
The outbreak of severe acute respiratory syndrome (SARS) in 2002–2003 has had a significant impact worldwide. No effective prophylaxis or treatment for SARS is available up to now. Angiotensin-converting enzyme 2 (ACE2) is the cellular receptor for SARS-associated coronavirus (SARS-CoV). By expressing a U6 promoter-driven small interfering RNA containing sequences homologous to part of ACE2 mRNA, we successfully silenced ACE2 expression in Vero E6 cells. By detecting negative strand SARS-CoV RNA and measuring RNA copy numbers of SARS-CoV by real-time reverse transcription polymerase chain reaction (RT-PCR), we demonstrated that SARS-CoV infection was reduced in the ACE2-silenced cell lines. These findings support the involvement of ACE2 in SARS-CoV infections and provide a basis for further studies on potential use of siRNA targeting ACE2 as a preventive or therapeutic strategy for SARS.
Keywords: Reverse Transcription Polymerase Chain Reaction, Severe Acute Respiratory Syndrome, Severe Acute Respiratory Syndrome, Quantitative Reverse Transcription Polymerase Chain Reaction, Severe Acute Respiratory Syndrome Patient
Introduction
The outbreak of severe acute respiratory syndrome (SARS) in 2002–2003 affected over 8,000 individuals and was responsible for almost 800 deaths (WHO, 2004). It also caused significant psychological and economic impacts worldwide [1, 2]. Although the SARS epidemic was stopped in mid-2003, the potential threat of SARS will remain high as long as no effective prevention or treatment for SARS is available.
Spike proteins of SARS-associated coronaviruses (SARS-CoV) interact with cellular receptors to mediate the infection of their target cells [3]. By using immunoprecipitation, Li et al. identified a metallopeptidase, angiotensin-converting enzyme 2 (ACE2), as the cellular receptor for SARS-CoV [4]. ACE2 is expressed mainly in the heart and kidneys but is also found in the bronchi and lungs [5, 6], where the major pathology of SARS resides.
RNA interference (RNAi) is a cellular process wherein short, double-stranded RNAs called small interfering RNAs (siRNAs) activate ribonucleases to target homologous mRNA [7, 8]. It has been used as a powerful tool for gene silencing. RNAi is also regarded as an antiviral strategy used by plants and probably other organisms [9]. Moreover, applications of RNAi technologies in the treatment or prevention of infectious diseases are being tested extensively.
The African green monkey kidney cell line Vero E6 expresses ACE2 and permits the replication of SARS-CoV [10]. In this study, we tested the hypothesis that RNAi technology can effectively eliminate ACE2, the cellular receptor for SARS-CoV, from Vero E6 cells and block SARS-CoV infections in these cells.
Material and methods
Plasmids
Plasmids generating small hairpin RNAs that target ACE2 mRNA were constructed using the vector pSilencer 2.1-U6 neo (Ambion, Austin, TX, USA). The mRNA of ACE2 (accession no. NM021804) was scanned for sequences of the AAN19TT pattern. These sequences were all analyzed by BLAST search of the GenBank database. Sequences with more than 16–17 contiguous base pairs of homology to other coding sequences were excluded. Two mutually complementary 64 oligonucleotides encoding small hairpin sequences targeting position 200–222 of human ACE2 mRNA (5′-GATCCGCCACGAAGCCGAAGACCTGTTCAAGAGA CAGGTCTTCGGCTTCGTGGTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAACCACGAAGCCGAAGACCTGTCTCTTGAACAGGTCTTCGGCTTCGTGGCG-3′) were synthesized, annealed, and ligated to the BamHI/HindIII sites of pSilencer 2.1-U6 neo to make pSilencer-ACE2. The inserted sequences were confirmed by direct sequencing.
Establishment of ACE2-silenced stable cell lines
The African green monkey kidney cell Vero E6 (CRL-1587) was purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in minimal essential medium with Earle’s balance salts (EMEM, JRH Biosciences, Lenexa, KS, USA) with 10% fetal bovine serum (FBS, Biological Industries, Kibbutz Bet Haemek, Israel). Vero E6 cells (3 × 105 cells/well) were seeded into 6-well plates in EMEM with 10% fetal calf serum 1 day before transfection in order to get a 70% confluence at transfection. pSilencer-ACE2 plasmids were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with a ratio of 4 ug DNA mix to 10 ul Lipofectamine 2000 per well, according to the manufacturer’s instructions. Transfection of the same plasmids was repeated once 48 h later. The cells were subcultured 24 h after the transfections, and then every 2–3 days in complete medium containing 2 mg/ml G418 (Sigma, St. Louis, MO, USA). The concentration of G418 was reduced to 1 mg/ml 14 days later. The cells were then diluted and single colonies were picked for further experiments.
SARS virus infection
The SARS-CoV strain TW1 [2], which was isolated from a confirmed SARS patient in the National Taiwan University Hospital, was used for infection. The stock virus was grown in Vero E6 cells, titered, and frozen in aliquots at −80°C. For quantitative analysis of SARS replication experiments, cells were grown to 80–90% confluence (~6 × 105 cells) in 12-well culture plates and inoculated with serially diluted SARS-CoV at multiplicity of infection (MOI) of 1, 10–1, 10–2, and 10–3. During the infection process, G418 was removed from the medium for the ACE2-silenced cells. For observation of cytopathic effects, 10–1 MOI of SARS viruses were inoculated to Vero E6 in 6-well plates. The virus were left in 200 ul EMEM with 2% FBS for 1 h for adsorption. Then, the cells were washed and re-fed with EMEM containing 10% FBS. Cytopathic effects were observed and RNA extraction was done at different time points after infection. All experiments associated with infectious SARS-CoV were carried out in a biosafety level 3 laboratory in the National Taiwan University.
Western blotting
Cell lysates were colleted by cell lysis buffer (10 mM Tris, 150 mM NaCl, 5 mM ethylenediaminetetraacetate, 1% NP40) premixed with protease inhibitor cocktail tablets (Roche, Penzberg, Germany), resolved by electrophoresis with 8% SDS-polyacrylamide gel, and transferred to a nitrocellulose membrane. Blocking was done with 5% skim milk. Goat anti-ACE2 antibody (AF933, R&D Systems, Minneapolis, MN, USA) in 1:100 dilution was used as the primary antibody and rabbit anti-goat horseradish peroxidase(HRP) (Chemicon, Temecula, CA, USA) in 1:5,000 dilution was used as the secondary antibody. The bands were visualized by using the enhanced chemiluminescence system (PerkinElmer, Boston, MA, USA). Quantitative analysis of chemiluminescence was done by using Kodak Image Station 2000R with Kodak 1D image analysis software (Rochester, NY, USA). ACE2 expressions were compared in different cells after normalization by β-actin expressions.
Flow cytometry analysis
Vero E6 and other cells were trypsinized and washed with 1% bovine serum albumin (BSA, Sigma, St. Louis, MO, USA) in phosphate-buffered saline (PBS). Aliquots of 106 cells were incubated in 100 ul PBS with or without 5 ug of ACE2 antibody for 60 min at 4°C. The cells were washed with 1% BSA/PBS and then incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat secondary antibody for 30 min at 4°C. The cells were then fixed with 2% formaldehyde and stored at 4°C until analysis. Data were acquired by the BD FACSCalibur system and analysis by Cell Quest (BD Bioscience, Franklin Lakes, NJ, USA).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Supernatants of cell cultures infected with the SARS-CoV were used for RNA extraction according to the mini spin protocol of the QIAamp virus RNA mini kit (Qiagen, Hilden, Germany). The extracted RNA was eluted in 50 µl RNase-free water, treated with deoxyribonuclease I (Life Technologies, Carlsbad, CA, USA) to digest genomic DNA, and stored at −80°C before use. Quantitative RT-PCR for SARS-CoV RNA was done by using the RealArt HPA-coronavirus LC RT-PCR Reagent (Artus Biotech, Hamburg, Germany) on a LightCycler instrument from Roche Diagnostics (Mannheim, Germany). Serial dilutions of the SARS-CoV stock were used as a standard.
Real-time RT-PCR was also done to quantify the amount of ACE2 mRNA. A primer pair that extends over two exons of ACE2 (1528-AAAGTGGTGGGAGATGAAGC and 1572-GTTTCATCATGGGGCACA) was used to amplify an amplicon of 63 nucleotides. A fluorophore and quencher dual-labeled probe (Human #77 from Exiqon, Vedbaek, Denmark) was used on the Roche LightCycler (Roche Diagnostics, Mannheim, Germany). GADPH mRNA was also amplified as an internal control and used for normalization with the primer pair CTCATGACCACAGTCCATGC (left) and CCCTGTTGCTGTAGCCAAAT (right).
Negative strand-specific RT-PCR
Negative strand-specific reverse transcription for SARS-CoV RNA was carried out with 0.1 ug of total RNA, 20 units of AMV reverse transcriptase (Roche Diagnostics, Indianapolis, IN, USA), and the sense primer (BNIoutS2: ATGAATTACCAAGTCAATGGTTAC) [11] at 42°C for 60 min. PCR with the primer pair ATGAATTACCAAGTCAATGGTTAC (BNIoutS2) and CATAACCAG TCGGTACAGCTA (BNIoutAs) [11] was then performed with universal cycle conditions (10 min at 95°C, 45 cycles of 10 s at 95°C, 10 s at 55°C, and 5 s at 72°C) on a LightCycler (Roche Diagnostics, Penzberg, Germany). The products were visualized by ethidium bromide staining after separating by 2–3% agarose gel electrophoresis. Again, GADPH mRNA was also amplified by PCR as an internal control with the primer pair CTCATGACCACAGTCCATGC (left) and CCCTGTTGCTGTAGCCAAAT (right).
Results
ACE2 expression was silenced in Vero E6 cells by siRNA
After two consecutive Lipofectamine transfections of pSilencer-ACE2, Vero E6 cells grew normally (Fig. 2b), with significantly reduced ACE2 expression. Western blotting showed different degrees of ACE2 expressions in various clones. Clones A4 and C4 both had >99% reductions in ACE2 expressions by Western blotting and were used for further experiments (Fig. 1a). Quantitative RT-PCR also revealed 96.2–96.7% reductions in ACE2 mRNA in both clones (Fig. 1b). Flow cytometry further confirmed that ACE2 expression was significantly reduced in clone A4 (Fig. 1c). Repeated Western blotting for ACE2 expression after removal of G418 proved that ACE2 of these cells were stably silenced (data not shown).
Fig. 2.
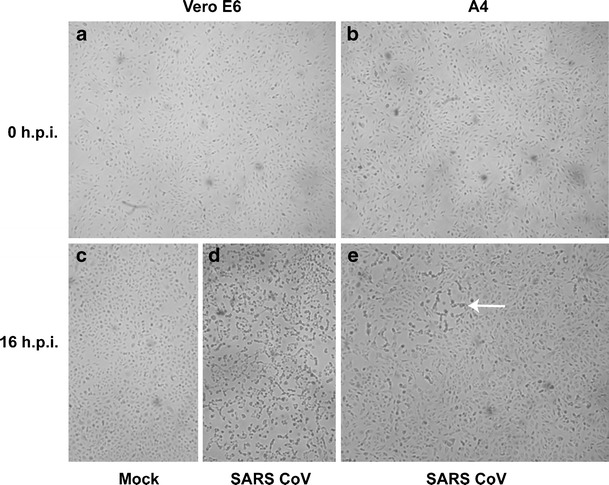
Inoculation of SARS-CoV resulted in typical cytopathic effects in Vero E6 but not A4 cells. Microscopic pictures of Vero E6 cells and its ACE2-silenced clone (A4) were taken before (a, b) and 16 h after (c, d, e) inoculation (h.p.i.) with SARS-CoV. Vero E6 cells inoculated with medium alone remained healthy and grew into a high density (c). At 16 h post-inoculation of SARS-CoV, almost all Vero E6 cells rounded up and died (d), while only a few foci of typical cytopathic effects (arrow in e) were shown in ACE2-silenced A4 cells (e)
Fig. 1.
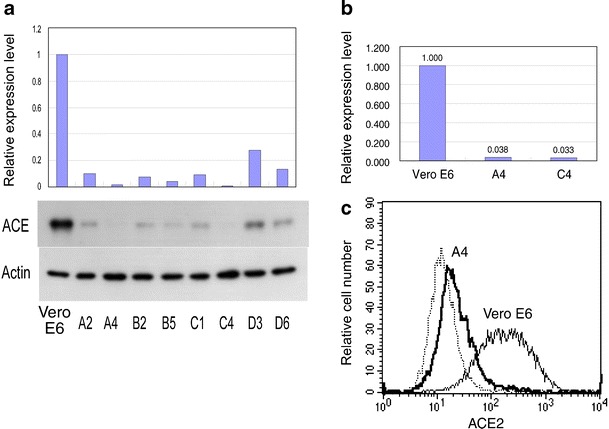
ACE2 was silenced by siRNA in Vero E6. a Western blotting of ACE2 in various clones of Vero E6 stably transfected with pSilencer-ACE2, which expressed siRNA targeting ACE2 (lower panel). Relative levels of ACE2 expression determined by quantitative analysis of chemiluminescence and normalized by β-actin expressions are shown in the upper panel. Clones A4 and C4 showed the lowest levels of ACE2 expression. b Quantitative RT-PCR of ACE2 mRNA. The numbers over the bars denote relative copy numbers of ACE2 mRNA normalized by GAPDH copy numbers. Clones A4 and C4 showed less than 4% of ACE2 mRNA left in comparison to the original Vero E6 cells. c Expressions of ACE2 in Vero E6 and clone A4 were determined by flow cytometry. The thin dotted line represents unstained control. ACE2 expression was hardly detectable in the majority of A4 cells
Replication of SARS-CoV was blocked in ACE2-silenced cells
Infections of SARS-CoV were done for Vero E6, A4, and C4 cells in a parallel fashion. Figure 2a and b show morphology of Vero E6 and A4 cells, respectively. Vero E6 showed good susceptibility to SARS-CoV. Almost all Vero E6 cells rounded up and died 16 h after inoculation (Fig. 2d). On the other hand, A4 and C4 cells remained grossly healthy at 16 h post-inoculation, except for a few foci of typical cytopathic effects (Fig. 2e).
By using a negative strand RNA-specific primer, the cDNA of the negative strand RNA of SARS-CoV RNA was synthesized and amplified. A product of 189 base pairs in size was detected in the Vero E6 but not in A4 or C4 cells (Fig. 3). SARS-CoV is a positive-stranded RNA virus. Replication of SARS-CoV required synthesis of a negative strand of SARS-CoV. Inability to detect a negative strand RNA in ACE2-silenced A4 and C4 cells confirmed the lack of SARS-CoV replication in these cells.
Fig. 3.
Negative strand of SARS-CoV RNA was detected in Vero E6 cells but not in ACE2-silenced A4 or C4 cells. RT-PCR using negative strand SARS-CoV RNA-specific primer for was done at 0 and 24 h after inoculation for Vero E6, A4, and C4 cells. A positive result for negative strand RNA of SARS-CoV was shown for Vero E6 cells at 24 h after inoculation. A4 and C4 cells whose ACE2 had been silenced showed no negative strand of SARS-CoV. GAPDH was amplified as an internal control
Production of SARS-CoV was compared between Vero E6 and A4 cells by determining SARS-CoV RNA copy numbers with qRT-PCR at various time points after inoculation (Fig. 4). With the inoculation dose of 1 MOI, SARS-CoV viral loads increased robustly to the same plateau in both Vero E6 and A4 cells. (Fig. 4a). When the inoculation dose decreased, the velocity of viral amplification also decreased. At inoculation doses of 10–1 or 10–2 MOI, SARS-CoV replication was significantly delayed in A4 cells. Lower viral loads were demonstrated in A4 than in Vero E6 at 24–48 h post-infection. However, the viral loads reached comparable levels in both cells at 72 h post-infection (Fig. 4b,c). When the inoculation dose was further decreased to 10–3 MOI, the virus did not replicate in A4 cells up to 72 h post-infection (Fig. 4d). The experiment was repeated twice with varied viral load readout but the same trends. The results shown in Fig. 4 were representative of the two independent experiments.
Fig. 4.
Replication of SARS-CoV was blocked in ACE2-silenced A4 cells at low inoculation doses. Viral loads of SARS-CoV were determined by real-time RT-PCR in Vero E6 and A4 cells with different inoculation doses at different time points. a At the inoculation dose of 1 MOI, no difference in viral loads was noted between Vero E6 and A4 cells at various time points. The velocities of SARS-CoV replication were significantly lower in A4 cells than in Vero E6 with the inoculation doses of 10–1 MOI (b) and 10–2 MOI (c). When the inoculation dose was decreased to 10–3 MOI, no increase in viral load was noted in A4 cells, while viral load increased to normal levels at 72 h in Vero E6 (d)
Discussion
The discovery of RNAi, together with development of methods for their generation and delivery, has led to potential application of RNAi as a novel antiviral approach. Reports regarding inhibition of SARS-CoV by RNAi are accumulating [12–21] and reviewed elsewhere [22, 23]. Using viral receptors or coreceptors as targets in inhibiting viral replication has also been tested for certain viruses, such as human immunodeficiency virus (HIV) [24, 25]. To our knowledge, however, this is the first study to test the feasibility of blocking SARS-CoV infections by siRNA targeting its cellular receptor, ACE2.
In several earlier studies testing the susceptibility of various permanent and primary eukaryotic cell lines to SARS-CoV, ACE2 mRNA could be detected in all of the susceptible cell lines and its abundance correlated with SARS-CoV susceptibility [26–28]. On the other hand, refractory cell lines were made permissive to SARS-CoV by the exogenous expression of ACE2 [29]. Our study extended these observations by showing that SARS-CoV susceptibility was reduced in Vero E6 cells as the ACE2 mRNA was reduced by siRNA.
Although successful silencing of ACE2 and reduction of susceptibility to SARS-CoV were shown in the current study, obstacles exist in clinical usage of RNAi-mediated ACE2 silencing as a preventive or therapeutic modality for SARS-CoV infections. The blocking effects of ACE2 silencing appeared to be modest. In our experiments with an inoculation dose of 10–1 to 10–2 MOI, viral loads reached nearly normal levels 72 h post-inoculation (Fig. 4b,c), suggesting as low as 4% of ACE2 expression (Fig. 1) is sufficient to support entry of SARS-CoV particles adequate to produce equivalent viral loads at 72 h. The presence of some other cellular receptor(s) for SARS-CoV is another possible explanation for the failure in blocking SARS-CoV replication at higher inoculations. CD209L was reported to be a minor receptor for SARS-CoV and has proved the possibility [30].
The efficacy of ACE2 silencing could theoretically be enhanced by choosing different targets, or using a “multiple-hit” approach by incorporating several siRNAs targeting various RNA sequences of SARS-CoV and its receptor(s) at the same time might be useful. Synergistic effects of more than one siRNA targeting different genes of SARS-CoV have been observed [31]. Besides, a recent study showed that interleukin (IL)-4 and interferon (IFN)-gamma can downregulate ACE2 expression on the cell surface and inhibit SARS-CoV replication [32]. “Cocktail” approaches by coadministration of RNAi and non-RNAi strategies to suppress ACE2 expression might maximize clinical effectiveness and hence have potential therapeutic applications.
Another concern about translating the current results to a clinically relevant context is that ACE2 was reported to be an essential regulator of cardiac function [33, 34]. ACE2 knockout mice showed cardiac dysfunction [34]. Selective elimination of ACE2 in organs vulnerable to SARS-CoV has never been tested in vivo and could possibly lead to unexpected consequences.
In summary, through siRNA technology, we have specifically silenced ACE2, the cellular receptor for SARS-CoV, in Vero E6 cells. Replication of SARS-CoV was reduced in these ACE2-silenced cells. The hypothesis that eliminating ACE2 can reduce or block SARS-CoV infections was proved to be true. However, considering the modest effect observed and concerns raised above, applying siRNA technology for SARS prevention or therapy requires further efforts.
Acknowledgments
This study was supported by research grant from the National Taiwan University Hospital (NTUH94A16). The funding source had no involvement in study design; collection, analysis, and interpretation of data; the writing of the report; and the decision to submit the paper for publication.
Conflict of interest statement
All authors declare no conflict of interest.
Footnotes
Part of the results was presented at the 184th Scientific Meeting of the Taiwan Pediatric Association (Taipei, 2006) and the 7th Annual Meeting of the Infectious Disease Society of Taiwan (Taipei, 2006).
References
- 1.Groneberg DA, Poutanen SM, Low DE, Lode H, Welte T, Zabel P. Treatment and vaccines for severe acute respiratory syndrome. Lancet Infect Dis. 2005;5:147–155. doi: 10.1016/S1473-3099(05)01307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsueh PR, Hsiao CH, Yeh SH, et al. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg Infect Dis. 2003;9:1163–1167. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donoghue M, Hsieh F, Baronas E, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 6.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/S0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 12.Li T, Zhang Y, Fu L, et al. siRNA targeting the leader sequence of SARS-CoV inhibits virus replication. Gene Ther. 2005;12:751–761. doi: 10.1038/sj.gt.3302479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin ZL, Zhao P, Zhang XL, et al. Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293T cells. Biochem Biophys Res Commun. 2004;324:1186–1193. doi: 10.1016/j.bbrc.2004.09.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Li T, Fu L, et al. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Shi Y, Yang DH, Xiong J, Jia J, Huang B, Jin YX. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 2005;15:193–200. doi: 10.1038/sj.cr.7290286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin ZL, Zhao P, Cao MM, Qi ZT. siRNAs targeting terminal sequences of the SARS-associated coronavirus membrane gene inhibit M protein expression through degradation of M mRNA. J Virol Methods. 2007;145:146–154. doi: 10.1016/j.jviromet.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Ren L, Zhao X, et al. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni B, Shi X, Li Y, Gao W, Wang X, Wu Y. Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA. Antivir Ther. 2005;10:527–533. [PubMed] [Google Scholar]
- 19.Akerstrom S, Mirazimi A, Tan YJ. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antiviral Res. 2007;73:219–227. doi: 10.1016/j.antiviral.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li BJ, Tang Q, Cheng D, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu CJ, Chan YL. Antiviral applications of RNAi for coronavirus. Expert Opin Investig Drugs. 2006;15:89–97. doi: 10.1517/13543784.15.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang Z, Babiuk LA, Hu J. Therapeutic and prophylactic potential of small interfering RNAs against severe acute respiratory syndrome: progress to date. BioDrugs. 2007;21:9–15. doi: 10.2165/00063030-200721010-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson J, Akkina R. CXCR4 and CCR5 shRNA transgenic CD34+ cell derived macrophages are functionally normal and resist HIV-1 infection. Retrovirology. 2005;2:53. doi: 10.1186/1742-4690-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novina CD, Murray MF, Dykxhoorn DM, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 26.Hattermann K, Muller MA, Nitsche A, Wendt S, Donoso Mantke O, Niedrig M. Susceptibility of different eukaryotic cell lines to SARS-coronavirus. Arch Virol. 2005;150:1023–1031. doi: 10.1007/s00705-004-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann H, Geier M, Marzi A, et al. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem Biophys Res Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie Y, Wang P, Shi X, et al. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem Biophys Res Commun. 2004;321:994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossel EC, Huang C, Narayanan K, Makino S, Tesh RB, Peters CJ. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J Virol. 2005;79:3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffers SA, Tusell SM, Gillim-Ross L, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He ML, Zheng BJ, Chen Y, et al. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett. 2006;580:2414–2420. doi: 10.1016/j.febslet.2006.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lang A, Osterhaus AD, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulter AB, Goddard MJ, Allen JC, Clark KL. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004;2:19. doi: 10.1186/1741-7015-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crackower MA, Sarao R, Oudit GY, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]


