Abstract
The aim of this was to investigate the relationship between single-nucleotide polymorphisms (SNPs) in the OAS1 gene and the susceptibility to chronic hepatitis C virus (HCV) infection in a population from the Liaoning Province of China. High resolution melt (HRM)-PCR analysis was conducted to examine three OAS1 SNPs: rs2660 G/A, rs10774671 G/A, and rs3741981 G/A in 298 chronic HCV-infected patients and in 305 healthy controls and to identify a relationship between SNP genotype and susceptibility to chronic HCV infection using a case–control study design. These three OAS1 SNPs were in strong linkage disequilibrium (rs2660 vs. rs10774671: |D’|=1.000, r 2 =1.000; rs2660/rs10774671 vs. rs3741981: |D’|=0.938, r 2 =0.569). The frequency of AG + GG genotypes in both rs2660 and rs10774671 and the AA + AG genotype in rs3741981 was significantly higher among chronic HCV-infected patients than among control (P < 0.001); the A allele in all three SNPs was found more frequently in the chronic HCV-infected group than in the control group (rs2660 and rs10774671: P = 0.02; rs3741981: P < 0.001). Moreover, individuals carrying the A allele in these SNPs exhibited an increased risk for chronic HCV infection (rs2660 and rs10774671: OR = 1.356 [1.051–1.749]; rs3741981: 1.363 [1.085–1.712]). The haplotype created by the G allele at both rs2660 and rs10774671 and the A allele at rs3741981 increased the risk of chronic HCV infection by 3.394-fold (95 % CI 1.406–8.201). Our results identify OAS1 SNP rs2660, rs10774671, and rs3741981 as genetic risk factors for chronic HCV infection. Polymorphisms of the OAS1 gene might affect the susceptibility to chronic infection with HCV.
Keywords: 2′,5′-Oligoadenylate synthetase gene; Chronic hepatitis C; High resolution melt; Single-nucleotide polymorphism; Haplotype
Introduction
Hepatitis C virus (HCV) infects 170 million patients worldwide and is one of the primary contributors to liver disease. Pegylated interferon alpha (Peg-IFN-α), in combination with ribavirin, is currently the standard-of-care for chronic HCV infection. The progression of disease following chronic HCV infection differs among individuals. This suggests that host genetic factors, such as single-nucleotide polymorphisms (SNPs), affect the course of chronic HCV infection.
2′,5′-Oligoadenylate synthetase (OAS) was among the first IFN-induced enzymes to be identified. The OAS/RNase L system plays a role in the innate immune response to viral infections. Several studies have identified SNPs in the OAS1 gene that influences the antiviral effects of IFNs to severe acute respiratory syndrome (SARS) or to West Nile virus infection. Both West Nile virus and HCV belong to the Flaviviridae. Therefore, we hypothesized that these same SNPs would affect the course of chronic HCV infection.
We used high resolution melt (HRM)-PCR analysis to evaluate whether OAS1 gene SNPs identified from a population in the Liaoning Province of China are associated with susceptibility to chronic HCV infection. From a group of 298 HCV cases and 305 healthy controls, we examined three OAS1 SNPs: a nonsynonymous A/G SNP in the sixth exon that corresponds to an Arg397Gly substitution (rs2660), an A/G SNP at the splice-acceptor site of exon 7 (rs10774671), and a nonsynonymous A/G SNP in the third exon that corresponds to Ser162Gly (rs3741981).
Materials and methods
Study population
Chronic HCV-infected Chinese patients (n = 298; 185 men, 113 women; mean age 56.92 ± 12.46 years) who consulted the First Hospital of China Medical University between November 2009 and April 2011 were selected for this study. Eligible patients tested positive for HCV antibody (Abbott Laboratories, USA) and had detectable (>5.0 × 102 IU/ml) serum HCV RNA as measured indirectly by real-time reverse-transcriptase PCR (QIAGEN, Germany). Patients positive for other hepatitis viral infection(s) were excluded from the study. The study was approved by the Institutional Ethics Committee of China Medical University, and all procedures followed institutional guidelines.
Healthy controls (n = 305; 210 men, 95 women; mean age: 57.63 ± 11.18 years) having had no history of hepatitis were selected among physical examination cases at the First Hospital of China Medical University between November 2009 and April 2011. The mean ages and gender ratios were not significantly different between the two groups (P = 0.99 and P = 0.21, respectively).
DNA isolation and genotyping
Genomic DNA was extracted from 100 μl of whole blood using the QIAamp DNA Micro Kit (QIAGEN), according to the manufacturer’s instructions. Extracted DNA was dissolved to 30 ng/μl (Thermo Fisher Scientific, USA) and was stored at −80 °C.
HRM-PCR (Roche Applied Science, Germany) was used for SNP genotyping. Samples were spiked with wild-type DNA as a standard [1]. Primers with annealing temperatures in the range of 55–60 °C that flanked short (100–150 bp) sequences were designed using Primer 5 (Table 1). Real-time PCR cycling and HRM analysis were conducted using Light Cycler® 480 System (Roche).
Table 1.
PCR primers and amplicons
| Polymorphism | Direction | Sequence | Amplicon size (bp) | Annealing temperature(°C) |
|---|---|---|---|---|
| rs2660 | Forward | gCACCATCCTCTgAATgCC | 149 | 59.2 |
| Reverse | AgACCTgggTTCTgTCCTg | 59.2 | ||
| rs10774671 | Forward | TCACAgTgTCTACCgTAAATg | 139 | 56.0 |
| Reverse | TTTCTgATACCTCCTgggATCg | 60.0 | ||
| rs3741981 | Forward | CTgACACCTAAgTTgTAgAT | 121 | 53.7 |
| Reverse | AACTCgCCCTCTTTCTgC | 57.3 |
PCR components included Master Mix (10 μl; Roche) containing FastStart Taq DNA polymerase reaction buffer, dNTPs, and HRM dye; 10 μM each forward and reverse primers (0.4 μl; Sangon Biotech, China), 25 nM MgCl2 (Roche), and 1.6 μl rs2660/rs3741981 or 2.0 μl rs10774671. Wild-type and mutant homozygotes were distinguished by spiking samples with a known genotype sequence before PCR. Unknown genomic DNA (30 ng) was used as template, along with an additional 6.0 ng (rs2660 and rs10774671) or 3.0 ng (rs3741981) of known wild-type DNA (obtained by sequencing previously). H2O (Roche) was added to bring the final reaction volumes to 20 μl.
PCRs were conducted in 96-well plates in 25-μl volumes using the following touchdown PCR cycling and HRM conditions: initiation with a 5-min hold at 95 °C, 40 cycles of 95 °C for 10 s, touchdown cycling (decreasing 1 °C/cycle), annealing in the range of 65–55 °C (rs2660) or 63–53 °C (rs10774671) or 60–50 °C (rs3741981) for 10 s, and 72 °C for 20 s. Following amplification, samples were heated to 95 °C for 30 s and then were cooled to 40 °C for 1 min to encourage heteroduplex formation. HRM curve data were obtained by melting over the range 65–95 °C at a rate of 25 data acquisitions per 1 °C. Results were analyzed using Light Cycler® 480 Gene Scanning software. 40 samples selected at random were verified by direct sequencing on an ABI7000 sequence detection system (Applied Biosystems, USA).
Statistical analysis
Data were analyzed using SPSS 17.0. Hardy–Weinberg equilibrium, linkage disequilibrium, and resultant haplotypes were examined using Haploview 4.0. Genotype and allele frequencies of the OAS1 gene in the healthy group and in the chronic HCV-infected group were analyzed using the chi-squared test. The relationship between polymorphisms and the risk to HCV were analyzed using the odds ratio (OR) and 95 % confidence intervals (CI). Statistical significance was set at P < 0.05.
Results
HRM analysis of OAS1 gene SNPs
HRM-PCR was used to genotype all samples for the OAS1 gene by spiking with the wild-type DNA standard (Fig. 1). Genotype results obtained using HRM or direct sequencing were consistent with each other.
Fig. 1.
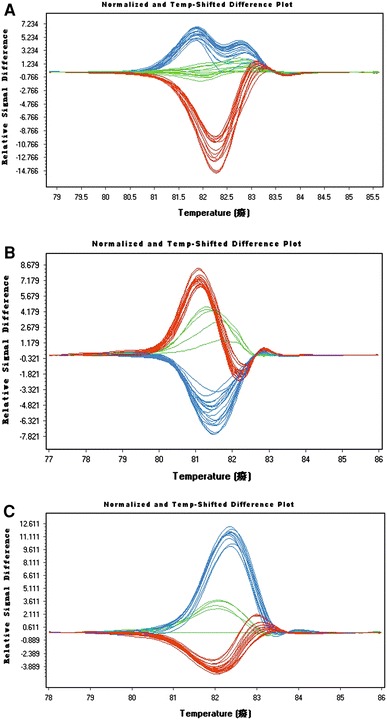
Genotyping results using HRM-PCR of OAS1 SNPs. a rs2660: red AA genotype, blue GA genotype, green GG genotype; b rs10774671: red GG genotype, blue GA genotype, green AA genotype; c rs3741981: red GG genotype, blue GA genotype, green AA genotype (color figure online)
OAS1 gene SNPs
The genotype and allele frequencies of the three OAS1 SNPs were distributed according to Hardy–Weinberg equilibrium in both the control and chronic HCV-infected groups (Table 2).
Table 2.
Genotype and allele frequencies of the OAS1 gene in the HCV-infected and control groups
| Patients (n = 298) | Controls (n = 305) | χ2 | P | OR (95 % CI) | |
|---|---|---|---|---|---|
| rs2660 | |||||
| Genotype | |||||
| AA | 174 (0.583) | 145 (0.475) | 7.12 | <0.001 | 1.548 |
| AG + GG | 104 (0.350) + 20 (0.067) | 136 (0.446) + 24 (0.077) | (1.123–2.134) | ||
| Allele | |||||
| A | 452 (0.758) | 426 (0.698) | 5.49 | 0.019 | 1.356 |
| G | 144 (0.242) | 184 (0.302) | (1.051–1.749) | ||
| rs10774671 | |||||
| Genotype | |||||
| AA | 174 (0.583) | 145 (0.475) | 7.12 | <0.001 | 1.548 |
| AG + GG | 104 (0.350) + 20 (0.067) | 136 (0.446) + 24 (0.077) | (1.123–2.134) | ||
| Allele | |||||
| A | 452 (0.758) | 426 (0.698) | 5.49 | 0.019 | 1.356 |
| G | 144 (0.242) | 184 (0.302) | (1.015–1.749) | ||
| rs3741981 | |||||
| Genotype | |||||
| AA + AG | 100 (0.336) + 157(0.527) | 94 (0.308) + 131(0.431) | 14.62 | <0.001 | 2.229 |
| GG | 41 (0.138) | 80 (0.261) | (1.478–3.362) | ||
| Allele | |||||
| A | 357 (0.599) | 319 (0.523) | 7.08 | <0.001 | 1.363 |
| G | 239 (0.401) | 291 (0.477) | (1.085–1.712) | ||
In the control population, the distribution frequencies of the AA, AG, and GG genotypes and of the A and G alleles at rs2660 were 47.5, 44.6, 7.9, 69.8, and 30.2 %, respectively. The genotype and allele frequencies at rs10774671 were the same as at rs2660. The frequencies at rs3741981 were 30.8, 43.1, 26.1, 52.3, and 47.7 %, respectively.
In terms of OAS1 gene SNP genotypes and allele frequencies between 298 chronic HCV-infected cases and 305 controls, we observed a higher frequency of the AA genotype at rs2660 in chronic HCV-infected patients than in controls (χ2 = 7.119, P < 0.001, OR = 1.548, 95 % CI 1.123–2.134). The allele frequency of the A allele at rs2660 was significantly higher in chronic HCV-infected patients than in controls (χ2 = 5.486, P = 0.019, OR = 1.356, 95 % CI 1.015–1.749). The genotype and allele frequencies at rs10774671 were the same as those at rs2660. The frequencies of the A-allele-positive genotypes (AA and GA) and of the A allele at rs3741981 were greater in the chronic HCV-infected group than in controls (AA and GA genotypes: χ2 = 14.615, P < 0.001, OR = 2.229, 95 % CI 1.478–3.362; A-allele: χ2 = 7.076, P = 0.008, OR = 1.363, 95 % CI 1.085–1.712).
Because linkage disequilibrium within a disease locus may be stronger for haplotypes than for single alleles, we examined whether previously determined haplotypes of OAS1 were associated with chronic HCV infection. Linkage disequilibrium characteristics between rs2660 and rs10774671 in our samples were as follows: |D, |= 1.000, r 2 = 1.000. Between rs2660/rs10774671 and rs3741981, linkage disequilibrium characteristics were as follows: |D, |= 0.938, r 2 = 0.569. The haplotype GGA, created by the G allele at rs2660 and rs10774671 and by the A allele at rs3741981 (hap 4), appeared 3.368-fold more frequently in the chronic HCV-infected patient group than in controls (P = 0.007). The other three haplotype frequencies did not differ significantly between the two groups (Table 3). The OR was calculated for hap4 versus the other three haplotypes using two-way contingency table analysis. Subjects carrying this haplotype had a 4.086-fold increased risk of chronic HCV infection (95 % CI 1.479–11.289).
Table 3.
Haplotype frequencies constructed with 3 OAS1 SNP in the normal group and hepatitis C group
| Haplotype (rs2660-rs10774671-rs3741981) | Control | Patients | χ2 | P | |
|---|---|---|---|---|---|
| Hap1 | A-A-A | 0.515 | 0.470 | 1.219 | 0.270 |
| Hap2 | G-G-G | 0.351 | 0.336 | 0.155 | 0.693 |
| Hap3 | A-A-G | 0.115 | 0.130 | 0.364 | 0.546 |
| Hap4 | G-G-A | 0.019 | 0.064 | 7.372 | 0.007 |
Discussion
The OASs are a family of IFN-induced enzymes that comprise the first IFN-induced enzymes to be identified. Four OAS genes, mapped to chromosome 12, have been described in humans: OAS1, OAS2, OAS3, and OASL (OAS-like) [2–5]. OAS enzymes are involved in the antiviral effect of IFN via the OAS/RNaseL pathway. Following exposure to novel viruses, IFN-I is one of the first cytokines released from immune-competent cells. IFN-I subsequently initiates an innate antiviral response via the induction of numerous IFN-stimulated genes. Among these are the OAS genes, which encode the synthesis of oligoadenylates of general structure ppp(A2′p9)nA (2′5′A); 2′5′A requires double-stranded (ds) RNA structures, such as viral genomes, to become activated. The activated enzyme then catalyzes the polymerization of ATP into 2′,5′-linked oligoadenylates (2′5′A). These, in turn, bind to and activate latent RNaseL, which degrades viral genetic material and inhibits protein synthesis [4–6].
Among healthy controls, the major OAS1 genotype for rs2660 and rs10774671 was AA, and the minor allele was A (Table 2). These results are consistent with those reported previously by Chen Lu-biao [7]. The major genotype for rs2660 and rs10774671 was AG, and the minor allele was A. This SNP distribution has not been reported previously; however, the data are consistent with those of the Chinese population in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP). The frequency distributions of these three sites in the Chinese population differ from those of Caucasian [8], Japanese [9], or Vietnamese [10] populations. Therefore, the genotypes and allele frequencies of these SNPs vary by race.
Knapp [11] reported previously that rs2660 was associated with chronic HCV infection. Hamano [10] and He [12] reported that the SNPs at rs2660 and rs3741981 were associated with SARS. Lim [13] suggested that the A allele at re10774671 was a risk factor for initial infection with West Nile Virus in humans. Fedetz [8] detected a link between the OAS1 gene SNPs rs10774671 and rs3741981 and susceptibility to multiple sclerosis. However, relationships to type I diabetes were inconsistent [14–18] until now.
We observed an association between rs2660/rs10774671 and chronic HCV infection; specifically, the AA genotype and the A allele both appeared more frequently among patients than among healthy controls. The SNP at rs2660 corresponds to the amino acid substitution, Arg397Gly. Although these two SNPs are in complete linkage disequilibrium (|D, |= 1.000, r 2 = 1.000), rs10774671 may be a more functional SNP because it is positioned at the last nucleotide of intron 5 in OAS1 and serves as a splice-acceptor site for exon 7. The A allele is predicted to abrogate splicing at this site. Splicing occurs further downstream, resulting in two mRNA forms, designated p48 and p52, which are associated with lower OAS enzymatic activity. The G allele at this site is predicted to allow splicing, giving rise to a p46 form with high enzymatic OAS activity. Heterozygotes exhibit intermediate OAS activity [10, 12, 13]. The p46 form more efficiently synthesizes 2′5′A, degrades viral genetic material, and inhibits protein synthesis. Hence, individuals carrying the G allele and GG + GA at this locus are less likely to become infected with chronic HCV.
In our study, the frequencies of the AA and AG genotypes at rs3741981 and the A allele were significantly higher in the chronic HCV-infected group compared with controls. Individuals with AA or AG genotypes or with the A allele at this locus were associated with an increased risk for chronic HCV infection. These data support a relationship between SNPs at rs3741981 and HCV susceptibility, with the AA and AG genotypes and the A allele being risk factors for chronic HCV infection. This is explained by the corresponding amino acid substitution at rs3741981. The OAS1 protein belongs to the dsDNA-binding protein family, which is characterized by one or more dsRNA-binding motifs (dsRBM) and by enzyme activation following binding between dsRBM and dsRNA [8, 19]. Substitutions of nearly any of the amino acids within the consensus dsRBM decreased dsRNA binding [20–22]. The A/G polymorphism in exon 3 of rs3741981 corresponds to the Ser162Gly substitution, which is located near the dsRNA-binding domain (amino acids 104–158) of OAS1. This decreases dsRNA binding and viral nucleic acid degradation. This may explain the observation that different individuals with different alleles exhibit different susceptibilities to chronic HCV infection.
The three OAS1 SNPs examined in this study were in strong linkage disequilibrium. Because of the complete linkage disequilibrium between rs2660 and rs10774671, only four haplotypes are created by these three alleles. Hap4, created by the G allele at rs2660 and rs10774671 and by the A allele at rs3741981, was more frequently detected in chronic HCV-infected patients than in controls. Haplotype analysis may identify relationships between disease-related polymorphisms more effectively than linkage disequilibrium analysis or single-locus analysis [23, 24]. We observed that individuals carrying hap4 had a significantly increased risk for chronic HCV infection.
There are many classical techniques for SNP genotyping, including denaturing high-performance liquid chromatography, single-strand conformation polymorphism, denatured gradient gel electrophoresis, and allele-specific oligonucleotide PCR. We performed HRM-PCR to identify SNPs. As a new generation of gene-scanning technology, HRM-PCR is a substantial improvement over other genotyping technologies in terms of its high-throughput, speed, simplicity, cost-effectiveness, probe-free design, and because it allows for closed-tube analysis. However, the detection of OAS1 gene SNPs using HRM-PCR has not been reported previously. We detected OAS1 gene SNPs in all samples we analyzed. Direct sequence verification demonstrated that the genotype results we obtained are reliable. Therefore, HRM-PCR is a simple and fast technique for detecting SNPs in the OAS1 gene.
HRM-PCR is an efficient, convenient, and low-cost method suitable for clinical analyses. We identified that OAS1 rs2660, rs10774671, and rs3741981 SNPs were associated with chronic HCV infection, and we demonstrated that these polymorphisms are associated with an increased risk of infection with HCV. The SNPs identified in this study using HRM-PCR may be applied as genetic protective factor for chronic HCV infection.
Acknowledgments
This work is supported by Liaoning provincial scientific research projects (L2010588). We appreciate the valuable comments from other members of our laboratories.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Yan Zhao, Email: equal521@163.com.
Hui Kang, Phone: +86-024-83282168, FAX: +86-024-83282678, Email: khui65@163.com.
Yangtang Ji, Email: jyt00@163.com.
Xin Chen, Email: cxin2005@126.com.
References
- 1.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 2.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/S1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 4.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL. Distinct antiviral roles for human 2′, 5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 6.Silverman RH. Viral encounters with 2′, 5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LB, Peng XM, Cao H, Zhang YF, Xu QH, Gao ZL. Relationship between SNP rs10774671 on OAS-1 gene and spontaneous HBeAg seroconversion in chronic HBV infection. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009;23:35–37. [PubMed] [Google Scholar]
- 8.Fedetz M, Matesanz F, Caro-Maldonado A, Fernandez O, Tamayo JA, Guerrero M, Delgado C, Lopez-Guerrero JA, Alcina A. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens. 2006;68:446–449. doi: 10.1111/j.1399-0039.2006.00694.x. [DOI] [PubMed] [Google Scholar]
- 9.Li CZ, Kato N, Chang JH, Muroyama R, Shao RX, Dharel N, Sermsathanasawadi R, Kawabe T, Omata M. Polymorphism of OAS-1 determines liver fibrosis progression in hepatitis C by reduced ability to inhibit viral replication. Liver Int. 2009;29:1413–1421. doi: 10.1111/j.1478-3231.2009.02061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamano E, Hijikata M, Itoyama S, Quy T, Phi NC, Long HT, Ha LD, Ban VV, Matsushita I, Yanai H, Kirikae F, Kirikae T, Kuratsuji T, Sasazuki T, Keicho N. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC, Hill AV, Thursz MR. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4:411–419. doi: 10.1038/sj.gene.6363984. [DOI] [PubMed] [Google Scholar]
- 12.He J, Feng D, de Vlas SJ, Wang H, Fontanet A, Zhang P, Plancoulaine S, Tang F, Zhan L, Yang H, Wang T, Richardus JH, Habbema JD, Cao W. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect Dis. 2006;6:106. doi: 10.1186/1471-2334-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, Stramer SL, Margolis LB, Murphy PM. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field LL, Bonnevie-Nielsen V, Pociot F, Lu S, Nielsen TB, Beck-Nielsen H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes. 2005;54:1588–1591. doi: 10.2337/diabetes.54.5.1588. [DOI] [PubMed] [Google Scholar]
- 15.Smyth DJ, Cooper JD, Lowe CE, Nutland S, Walker NM, Clayton DG, Todd JA. No evidence for association of OAS1 with type 1 diabetes in unaffected siblings or type 1 diabetic cases. Diabetes. 2006;55:1525–1528. doi: 10.2337/db05-1452. [DOI] [PubMed] [Google Scholar]
- 16.Tessier MC, Qu HQ, Frechette R, Bacot F, Grabs R, Taback SP, Lawson ML, Kirsch SE, Hudson TJ, Polychronakos C. Type 1 diabetes and the OAS gene cluster: association with splicing polymorphism or haplotype? J Med Genet. 2006;43:129–132. doi: 10.1136/jmg.2005.035212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howson JM, Walker NM, Smyth DJ, Todd JA, Type IDG.C. Analysis of 19 genes for association with type I diabetes in the Type I Diabetes Genetics Consortium families. Genes Immun. 2009;10(suppl 1):S74–S84. doi: 10.1038/gene.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu HQ, Polychronakos C, Type IDGC Reassessment of the type I diabetes association of the OAS1 locus. Genes Immun. 2009;10(suppl 1):S69–S73. doi: 10.1038/gene.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F. Variation in antiviral 2′, 5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 21.Green SR, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 22.Krovat BC, Jantsch MF. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J Biol Chem. 1996;271:28112–28119. doi: 10.1074/jbc.271.45.28112. [DOI] [PubMed] [Google Scholar]
- 23.Huang Q, Fu YX, Boerwinkle E. Comparison of strategies for selecting single nucleotide polymorphisms for case/control association studies. Hum Genet. 2003;113:253–257. doi: 10.1007/s00439-003-0965-x. [DOI] [PubMed] [Google Scholar]
- 24.Lautier C, El Mkadem SA, Renard E, Brun JF, Gris JC, Bringer J, Grigorescu F. Complex haplotypes of IRS2 gene are associated with severe obesity and reveal heterogeneity in the effect of Gly1057Asp mutation. Hum Genet. 2003;113:34–43. doi: 10.1007/s00439-003-0935-3. [DOI] [PubMed] [Google Scholar]


