Abstract
Staphylococcus aureus is an important etiological agent responsible for healthcare-associated infections. In this study, the effect of flavonoids on the inhibition of S. aureus PriA (SaPriA), an essential helicase for DNA replication restart, which is critical for bacterial survival, was investigated. Using vanadate-sensitive colorimetric assay, the concentration of phosphate, from ATP hydrolysis by SaPriA, was decreased to 37 and 69 %, respectively, in the presence of 35 μM kaempferol and myricetin. The effect of quercetin, galangin, dihydromyricetin, and myricitrin was insignificant. From titration curve, IC50 of kaempferol for SaPriA was determined to be 22 ± 2 μM. Using fluorescence quenching, we identified that kaempferol can bind to SaPriA with K d of 9.1 ± 3.2 μM. To our knowledge, these preliminary results constituted the first study regarding that naturally occurring product such as flavonols kaempferol and myricetin can be potent inhibitors targeting PriA.
Keywords: Staphylococcus aureus, PriA helicase, Inhibitor, Flavonol, Kaempferol, Myricetin, Drug development
Introduction
DNA helicases are required in most stages of DNA metabolism, ranging from DNA replication, repair, and recombination [1, 2]. DNA helicases utilize energy via the hydrolysis of nucleoside triphosphate (NTP) to move along nucleic acid filaments with either a 5′–3′ or 3′–5′ direction and separate double-stranded DNA (dsDNA) into the complementary single-stranded DNA (ssDNA). Helicases were grouped into several superfamilies (SF) on the basis of the helicase motifs. Most SF1 and SF2 helicases have low activity in unwinding dsDNA when acting without their partner proteins and might need these accessory proteins to stimulate the helicase activity. In addition, helicases are also needed for the removal of nucleoproteins from DNA in many different stages of DNA metabolism and gene expression [1].
PriA is a SF2 helicase possessing ATPase and 3′–5′ helicase activities and is originally discovered as a major factor essential for phage ϕX174 replication [3]. PriA is a poor helicase when acting alone in vitro and might need other accessory proteins, such as PriB and SSB, to stimulate the helicase activity [4, 5]. The PriA-orientated replication restart primosome is a formidable enzymatic machine used in reactivation of stalled DNA replication [3]. Unlike the DnaA-directed primosome, which is always initiated at the unique oriC site for further assembly, the PriA-directed replication restart primosome does not assembled for specific DNA sequence but preferentially recognizes branched DNA structures [6–8]. The mechanisms of action of replication restart primosome in the Gram-negative Escherichia coli have been established [9–11]. In E. coli, the replication restart primosome consists of PriA helicase, PriB, PriC, DnaB helicase, DnaC, DnaT, and DnaG primase [12]. In the Gram-positive Bacillus subtilis, the DNA replication initiator protein PriA helicase has homolog of E. coli [13]. Nevertheless, PriB, PriC, DnaT, and DnaC proteins, essential components of the replication restart primosome, are not found in Gram-positive bacteria. Instead, DnaD, DnaB, and DnaI are found as essential factors for replication restart of the Gram-positive B. subtilis. On the basis of the function and activity, DnaI may be the Gram-positive functional counterpart of E. coli DnaC, a helicase loader protein [14].
Flavonoids are plant polyphenols [15], and many display unique biological activities which have been used as some pharmaceutical agents [16]. Flavonols belong to one of the six major subclasses of flavonoids, and others are flavones, flavanones, flavanols, anthocyanidins, and isoflavones. Some flavonols are known to have significant antioxidant [17], antiradical [18], and antibacterial activities [19–22]. Recently, few therapies are effective against the antibiotic-resistant pathogens, especially the six antibiotic-resistant ESKAPE pathogens [23, 24]. S. aureus, which is a Gram-positive pathogen, has a remarkable ability to develop antibiotic resistance. Although some advances in treatment and prevention are made, S. aureus still create serious problems to public health worldwide. It is thought that PriA is an essential initiator protein required for restart of DNA replication in the Gram-positive bacteria, inhibiting the activity of PriA will be detrimental to block bacterial growth and survival.
Materials and Methods
The gene SAAV1184, encoding SaPriA, was amplified by PCR using genomic DNA of Staphylococcus aureus subsp. aureus ED98 as template. The forward (5′-GGGAAGGATCCATGATAGCGAAAGTCA-3′) and the reverse (5′-CCATTCTCGAGCATCATCATCTGTGGA-3′) primers were designed, and BamHI and XhoI restriction sites into SaPriA were introduced. The PCR product was then ligated to the pET21b vector. SaPriA was purified by Ni2+-affinity chromatography (GE Healthcare Bio-Sciences) eluted with Buffer A (20 mM Tris–HCl, 250 mM imidazole, and 0.5 M NaCl, pH 7.9). After dialysis against Buffer B (20 mM HEPES and 100 mM NaCl, pH 7.0), the SaPriA solution was further purified by the Heparin HP column (GE Healthcare Bio-Sciences), eluted with a linear NaCl gradient from 0.1 to 1.0 M with Buffer B using the AKTA-FPLC system (GE Healthcare Bio-Sciences). SDS-PAGE was used to show the protein purity (Fig. 1). By the vanadate-sensitive colorimetric assay, the ATPase activity of SaPriA was detected: more the concentration of inorganic phosphate released by ATP hydrolysis, more the OD610 intensity. The binding of kaempferol to SaPriA was analyzed by the fluorescence emission spectra of SaPriA quenched by kaempferol.
Fig. 1.

SDS-PAGE of the purified SaPriA and molecular mass standards
Results
Myricetin is a flavonol, and has three hydroxyl substituents on the aromatic ring (Fig. 2). The inhibitory effect of myricetin on several helicases, such as the SARS coronavirus helicase [25], the replicative DnaB helicase [21, 26, 27], and RSF1010 RepA helicase [28], has been established. Given that the inhibitory effect of myricetin on PriA helicase is not known yet, we used myricetin as a potential inhibitor on SaPriA. Other flavonoids, namely, dihydromyricetin, myricitrin, quercetin, galangin, and kaempferol, were further used to test the structure–inhibition relationship for SaPriA. We analyzed the ATPase activity of SaPriA by detecting the concentration of inorganic phosphate released by ATP hydrolysis [29]. The concentration of phosphate from ATP hydrolysis by SaPriA was decreased to 37 and 69 %, respectively, in the presence of 35 μM kaempferol and myricetin (Fig. 3). The effect of quercetin, galangin, dihydromyricetin, and myricitrin was insignificant (Fig. 3). From titration curve, IC50 of kaempferol for SaPriA was determined to be 22 ± 2 μM (Fig. 4). To our knowledge, this is the first study regarding that naturally occurring product such as flavonols kaempferol and myricetin can be potent inhibitors targeting PriA.
Fig. 2.
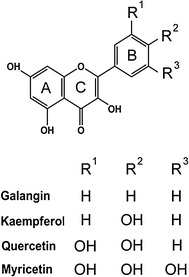
Molecular structure of myricetin, quercetin, kaempferol, and galangin
Fig. 3.
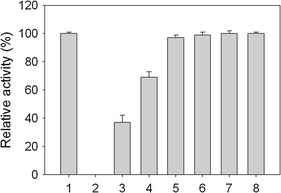
Inhibition of the ATPase activity of SaPriA. The ATPase activity of purified SaPriA was analyzed using the vanadate-sensitive colorimetric assay. The ATPase activity assay (2 mL of reaction volume) for SaPriA (2 μM) was analyzed in 20 mM HEPES (pH 7.0), 5 mM of MgCl2, 1 mM of ATP, and 35 μM of flavonoid for 2 h, and then the OD610 was analyzed. The reaction mixture with (lane 1) or without SaPriA (lane 2) was analyzed. The reaction mixture with SaPriA was analyzed in the presence of kaempferol (lane 3), myricetin (lane 4), dihydromyricetin (lane 5), galangin (lane 6), quercetin (lane 7), and myricitrin (lane 8)
Fig. 4.
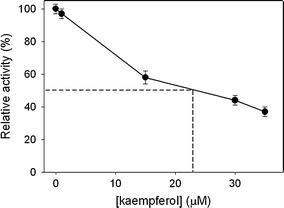
The titration curve of kaempferol for SaPriA. IC50 of kaempferol was determined to be 22 ± 2 μM using graphic analysis
To determine whether kaempferol can bind to SaPriA, the fluorescence emission spectra of SaPriA quenched by kaempferol are shown in Fig. 5. Quenching refers to the complex formation process that decreases the fluorescence intensity of the protein. As kaempferol (0–10 μM) was added into the SaPriA solution, the intrinsic fluorescence intensity of the protein was progressively decreased. The maximum λem of SaPriA was slightly shifted (~1.5 nm). Therefore, these results from the quenching of SaPriA fluorescence indicated the formation of a complex between kaempferol and SaPriA. In addition, the intrinsic fluorescence of SaPriA at 330 nm, which is excited at 280 nm, was significantly quenched by 55 % in the presence of 10 μM kaempferol. K d value of SaPriA bound to kaempferol determined from the titration curve (not shown) were 9.1 ± 3.2 μM. Thus, kaempferol can bind to SaPriA and then inhibit its ATPase activity.
Fig. 5.
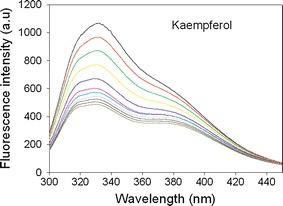
The fluorescence quenching of SaPriA by kaempferol. Compound concentrations, from the top down, are 0 to 10 μM. Fluorescence titration was performed using a spectrofluorimeter (Hitachi F-2700). An aliquot amount of kaempferol was added into the solution containing SaPriA (0.5 μM), 20 mM HEPES, and 100 mM NaCl at pH 7.0. The K d value was obtained by the equation: ΔF = ΔFmax − K d(ΔF/[compound]) (Enzyme Kinetics module of Sigma-Plot)
Discussion
S. aureus, a Gram-positive pathogen, exhibits a remarkable ability to develop antibiotic resistance [30]. Development of clinically useful small molecule antibiotics is highly needed to target S. aureus and other infections [30]. Considering that PriA-directed primosomes are required for bacterial DNA replication restart processes, PriA may be a suitable target for antibiotic development. In addition, PriA is not found in humans; hence, inhibitors based on PriA inhibition are potentially safe for human use. Flavonoids are plant polyphenols, and many are known to have antiradical, antiviral, antioxidant, and antibacterial activities. Some flavonoids are also served as the ATPase-inhibiting agents for competition with ATP binding of the protein, and thus, these flavonoid derivatives may be the lead compounds for developing therapeutic agents for fighting cancer [31]. Our laboratory is also currently screening some helicases from Hepatitis viruses using these flavonoids and the derivatives.
Conclusion
In this study, we analyzed the effects of the flavonoids, namely, myricetin, dihydromyricetin, myricitrin, quercetin, galangin, and kaempferol, on ATP hydrolysis ability of SaPriA. For the first time, our results demonstrated that naturally occurring products such as flavonols kaempferol and myricetin were capable of inhibiting the PriA activity. These flavonol compounds as well as their derivatives may be used as lead compounds in the development of new antibiotics that target S. aureus and other bacteria.
Acknowledgments
This research was supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 103-2320-B-040-018-MY2 to C.Y. Huang).
Abbreviations
- SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- Kd
The apparent dissociation constant
- Sa
Staphylococcus aureus
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 3.Masai H, Tanaka T, Kohda D. Stalled replication forks: making ends meet for recognition and stabilization. BioEssays. 2010;32:687–697. doi: 10.1002/bies.200900196. [DOI] [PubMed] [Google Scholar]
- 4.Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P. PriB stimulates PriA helicase via an interaction with single-stranded DNA. J Biol Chem. 2005;280:39693–39700. doi: 10.1074/jbc.M508521200. [DOI] [PubMed] [Google Scholar]
- 5.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Mizukoshi T, Sasaki K, Kohda D, Masai H. Escherichia coli PriA protein, two modes of DNA binding and activation of ATP hydrolysis. J Biol Chem. 2007;282:19917–19927. doi: 10.1074/jbc.M701848200. [DOI] [PubMed] [Google Scholar]
- 7.Mizukoshi T, Tanaka T, Arai K, Kohda D, Masai H. A critical role of the 3′ terminus of nascent DNA chains in recognition of stalled replication forks. J Biol Chem. 2003;278:42234–42239. doi: 10.1074/jbc.C300285200. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Mizukoshi T, Taniyama C, Kohda D, Arai K, Masai H. DNA binding of PriA protein requires cooperation of the N-terminal D-loop/arrested-fork binding and C-terminal helicase domains. J Biol Chem. 2002;277:38062–38071. doi: 10.1074/jbc.M204397200. [DOI] [PubMed] [Google Scholar]
- 9.Fujiyama S, Abe Y, Tani J, Urabe M, Sato K, Aramaki T, Katayama T, Ueda T. Structure and mechanism of the primosome protein DnaT-functional structures for homotrimerization, dissociation of ssDNA from the PriB∙ssDNA complex, and formation of the DnaT∙ssDNA complex. FEBS J. 2014;281:5356–5370. doi: 10.1111/febs.13080. [DOI] [PubMed] [Google Scholar]
- 10.Huang YH, Lin MJ, Huang CY. DnaT is a single-stranded DNA binding protein. Genes Cells. 2013;18:1007–1019. doi: 10.1111/gtc.12095. [DOI] [PubMed] [Google Scholar]
- 11.Huang YH, Huang CY. The N-terminal domain of DnaT, a primosomal DNA replication protein, is crucial for PriB binding and self-trimerization. Biochem Biophys Res Commun. 2013;442:147–152. doi: 10.1016/j.bbrc.2013.11.069. [DOI] [PubMed] [Google Scholar]
- 12.Huang YH, Huang CY. Structural insight into the DNA-binding mode of the primosomal proteins PriA, PriB, and DnaT. Biomed Res Int. 2014;2014:195162. doi: 10.1155/2014/195162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/S1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 14.Soultanas P. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 2002;30:966–974. doi: 10.1093/nar/30.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross JA, Kasum CM. Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 16.Teillet F, Boumendjel A, Boutonnat J, Ronot X. Flavonoids as RTK inhibitors and potential anticancer agents. Med Res Rev. 2008;28:715–745. doi: 10.1002/med.20122. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe KL, Liu RH. Structure-activity relationships of flavonoids in the cellular antioxidant activity assay. J Agric Food Chem. 2008;56:8404–8411. doi: 10.1021/jf8013074. [DOI] [PubMed] [Google Scholar]
- 18.Burda S, Oleszek W. Antioxidant and antiradical activities of flavonoids. J Agric Food Chem. 2001;49:2774–2779. doi: 10.1021/jf001413m. [DOI] [PubMed] [Google Scholar]
- 19.Daglia M. Polyphenols as antimicrobial agents. Curr Opin Biotechnol. 2012;23:174–181. doi: 10.1016/j.copbio.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Chirumbolo S. Plant polyphenolic compounds as potential antimicrobial drugs. J Med Microbiol. 2011;60:1562–1563. doi: 10.1099/jmm.0.032201-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen CC, Huang CY. Inhibition of Klebsiella pneumoniae DnaB helicase by the flavonol galangin. Protein J. 2011;30:59–65. doi: 10.1007/s10930-010-9302-0. [DOI] [PubMed] [Google Scholar]
- 22.Cushnie TP, Lamb AJ. Antimicrobial activity of flavonoids. Int J Antimicrob Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush K. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 25.Keum YS, Jeong YJ. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin HH, Huang CY. Characterization of flavonol inhibition of DnaB helicase: real-time monitoring, structural modeling, and proposed mechanism. J Biomed Biotechnol. 2012;2012:735368. doi: 10.1155/2012/735368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griep MA, Blood S, Larson MA, Koepsell SA, Hinrichs SH. Myricetin inhibits Escherichia coli DnaB helicase but not primase. Bioorg Med Chem. 2007;15:7203–7208. doi: 10.1016/j.bmc.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Ziegelin G, Schroder W, Frank J, Ayora S, Alonso JC, Lanka E, Saenger W. Flavones inhibit the hexameric replicative helicase RepA. Nucleic Acids Res. 2001;29:5058–5066. doi: 10.1093/nar/29.24.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-X. [DOI] [PubMed] [Google Scholar]
- 30.Koyama N, Inokoshi J, Tomoda H. Anti-infectious agents against MRSA. Molecules. 2012;18:204–224. doi: 10.3390/molecules18010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chahar MK, Sharma N, Dobhal MP, Joshi YC. Flavonoids: a versatile source of anticancer drugs. Pharmacogn Rev. 2011;5:1–12. doi: 10.4103/0973-7847.79093. [DOI] [PMC free article] [PubMed] [Google Scholar]


