Abstract
To reduce morbidity and mortality through integrated case management, a pilot study to detect respiratory viruses in patients with acute lower respiratory infections (ALRIs) was designed as part of a nationwide surveillance for this disease in Korea. The study population consisted of hospitalized patients under the age of 5 years with bronchiolitis, pneumonia, croup, or acute respiratory distress syndrome. A prospective 6-month study was performed. Two hundred and ninety-seven nasopharyngeal secretions were collected and multiplex reverse transcriptase polymerase chain reactions (RT-PCR)/polymerase chain reactions (PCR) were performed to detect respiratory viruses. If there were any positive RT-PCR/PCR results, viral cultures were proceeded for confirmation. Respiratory viruses were identified in 49.6% of 296 patients. The detection rates were as follows: respiratory syncytial virus (RSV) was the most commonly detected in 52.7% (87/165), human metapneumovirus (hMPV) in 15.8%, human corona virus (hCoV) in 5.5%, adenovirus in 9.7%, human bocavirus (hBoV) in 5.5%, parainfluenza virus (PIV) in 3.6%, rhinovirus (RV) in 4.2%, and the influenza virus in 3% of the patients with ALRIs. The consistent rate of positive results between RT-PCR and viral culture was 92% (105/114). Using our methods to detect viral causes seemed to be acceptable for the national surveillance of severe acute respiratory infections in infants and children.
Keywords: Influenza Virus, Respiratory Syncytial Virus, Reverse Transcriptase Polymerase Chain Reaction, Acute Respiratory Distress Syndrome, Respiratory Syncytial Virus Infection
Acute lower respiratory infections (ALRIs) has a major impact on health services and account for up to 50% of visits by children to health facilities. Many emerging viral infections causing ALRIs may not be detected until the middle of an outbreak. In 2006 spring, throughout almost every tertiary hospital in South Korea, certain patients were admitted for more than 1 month due to fatal respiratory infections and pediatricians could not determine the causal virus of the outbreak. Rapidly progressive interstitial pneumonia, subsequent pneumothorax, prolonged ventilator care with diagnosis of acute respiratory distress syndrome (ARDS), and multiple organ failures were typical disease progression. Early molecular viral studies on possible patients could be the first key to detecting causal viruses [1, 2], so a nationwide surveillance system to control the spreading of infections was designed by the Korea Centers for Disease Control and Prevention (KCDC).
This pilot study was conducted to establish a nationwide surveillance system of ALRIs in infants and children under 5 years of age in South Korea. Including classic respiratory viruses, such as the respiratory syncytial virus (RSV), parainfluenza virus (PIV), influenza virus, adenovirus, and rhinovirus (RV), and recently discovered viruses, e.g., human metapneumovirus (hMPV), human bocavirus (hBoV), and human corona viruses (hCoV)-OC 43, 229E were scanned by the multiplex reverse transcriptase polymerase chain reactions (RT-PCR)/polymerase chain reactions (PCR) method. In detecting causal respiratory viruses, this molecular tool has been known to be the most sensitive [3]. In the present study, the feasibility of establishing a hospital-based respiratory viral surveillance system and a prompt reporting system between tertiary hospitals and the KCDC were investigated.
ALRI patients under 5 years of age (n = 296) admitted in Severance Children’s Hospital, which is one of the largest tertiary hospitals in South Korea, were examined prospectively between November 2007 and April 2008 for 6 months. Case definitions of moderate to severe lower respiratory infections were determined according to the World Health Organization (WHO) recommended surveillance standards 2nd edition (1999). As part of the diagnosis, pneumonia, croup, bronchiolitis, and ARDS were included, which were diagnosed by the same physician through physical examination and chest X-rays. Samples were collected by nasopharyngeal aspiration with viral transport media prospectively and informed consent was obtained. This study was approved by the Institutional Review Board of Severance Hospital. Questionnaires asked about the onset and manifestation of symptoms, admission and duration of stay, sample collection, risk factors, e.g., parents smoking, prematurity, formula feeding, gastro-esophageal reflux, neurologic deficits, cardiovascular defects, initial diagnosis, radiographic findings, blood tests, and treatments. The information was provided by the same pediatrician and was entered into an Access 2007 database (Microsoft) by a person blinded to the viral results. Immunocompromised patients, such as patients receiving chemotherapy due to various cancers, those taking immunosuppressants, patients who were admitted to the neonatal intensive care unit, and patients with severe complex heart diseases, were excluded. All analyses were performed with GraphPad Prism version 5.00 for Windows (GraphPad Software Inc., San Diego, CA, USA). A P-value < 0.05 was considered to be significant.
Viral DNA or RNA was extracted with a QIAamp Viral Mini Kit (QIAGEN, Hilden, Germany), and One-step Multiplex PCR/RT-PCR reagents and the Labopass™ RV Detection kit (Cosmo GENETECH, Seoul, South Korea), which was developed by the KCDC influenza and respiratory virus team with Cosmo GENETECH, were used. Samples showing any positive results for adenovirus, influenza virus, PIV, and RSV by multiplex PCR methods underwent viral culture through R-Mix™ ReadyCells (Diagnostic Hybrids, Athens, US).
Among a total of 296 patients, 297 samples were collected. Of 297 samples, 147 samples (49.6%) were positive and, in total, 165 viruses were detected through this study. RSV was the most commonly detected in 52.7% (87/165), hMPV in 15.8% (26/165), adenovirus in 9.7% (16/165), hBoV in 5.5% (9/165), hCoV in 5.5% (9/165), RV in 4.2%(7/165), PIV in 3.6% (6/165), and the influenza virus in 3% (5/165) of the patients with ALRIs. The co-infection rate was 11.6%. The detection frequency of each virus in ALRIs with respect to demographic features and clinical manifestations are depicted in Table 1. The elapsed period of the initial presentation of symptoms and the time of sampling were not significantly different between the virus-positive samples and the virus-negative samples (data not shown). However, among the detected viruses, significant differences in the elapse of sampling was noted by one-way analysis of variance (ANOVA; P = 0.03). The sampling time was prominently different between adenovirus and hMPV (P < 0.01), but there was no significant difference between RSV and hMPV or hBo and hMPV (Fig. 1).
Table 1.
The clinical manifestations of acute lower respiratory infections (ALRIs) according to each of the viruses
| Adeno | hBo | PIV | RSV | hCoV | RV | Influenza | hMPV | |
|---|---|---|---|---|---|---|---|---|
| Number of patients (total = 296) | 16 | 9 | 6 | 87 | 9 | 7 | 5 | 26 |
| Median age (range) | 24 months (3–58) | 28 months (3–41) | 28 months (0–69) | 13 months (2–66) | 9 months (5–39) | 10 months (2–48) | 22 months (14–49) | 13 months (1–66) |
| Median hospital stay(range) | 4 days (4–50) | 3 days (2–7) | 4 days (2–14) | 4 days (1–68) | 4 days (5–39) | 3 days (2–48) | 5 days (4–14) | 4 days (1–66) |
| High fever (%)* | 81.8 | 63.6 | 73.3 | 68 | 42.8 | 85.7 | 100 | 84 |
| Coinfection (patients) | 4 | 6 | 2 | 12 | 1 | 3 | 1 | 12 |
| Diagnosis (patients) | ||||||||
| Pneumonia | 14 | 4 | 3 | 62 | 3 | 8 | 4 | 22 |
| Bronchiolitis | 1 | 3 | 0 | 21 | 2 | 4 | 1 | 3 |
| Croup | 0 | 2 | 3 | 3 | 4 | 0 | 0 | 1 |
| ARDS | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
*The proportion of patients who presented with high fever
Fig. 1.
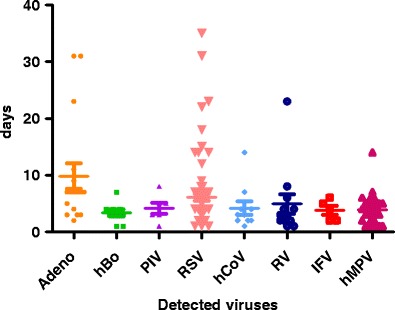
Elapsed time differences between initial symptom presentation and nasopharyngeal samplings in each of the viruses. The bars represent mean values with 95% confidence intervals
In the course of this study, RSV outbreak was observed from November to December such that RSV was detected in about 70% of cases enrolled in this period. There was no significant difference in the clinical outcomes, such as length of hospital stay, indices of respiratory difficulty, and oxygen therapy among groups of RSV infections (75 cases), RSV+hMPV coinfections (8 cases), and hMPV infections (13 cases).
Pneumonia was diagnosed in 75%, bronchiolitis in 17%, croup in 7%, and ARDS in 1% of patients. Among the patients, 15/296 (5.4%) needed oxygen therapy and 8/296 (2.7%) were treated with a ventilator. In ARDS patients (n = 4), only two cases were revealed as causal viruses, which were adenovirus and RSV. Two cases resulted in mortality in spite of meticulous treatments.
A total of 211 parents responded to the questionnaires, 144 patients were breast-fed (68.2%) for more than 1 month. The ages of 42 of 211 (20%) patients were less than 6 months old. In this group, breast-feeding did not affect the rate of virus detection (P = 0.23). Among formula-fed infants, there was no difference in terms of viral detection between the older than 6 months and younger than 6 months groups (P = 0.68). Of the parents, 106/211 smoked (50.2%). The respiratory difficulty index, items such as nasal flaring, tachypnea (RR > 40/min), or chest wall retractions, were significantly higher in the smoking group as determined by logistic regression analysis with other cofactors (RSV infection and asthma) (P = 0.04). ALRI patients with a history of prematurity (gestational age 26–36 weeks) were found in 12 patients (4%). Their median age was 19 months. One patient had received RSV monoclonal antibody. In this group, there were 6 of 12 virus-positive patients (50%), all of which were RSV, except for one case.
The positive samples in which a single virus was detected by multiplex RT-PCR/PCR were put into viral cultures. RSV, adenovirus, PIV 1,2,3, and influenza A and B were isolated using this method. Viral cultures were performed in 114 samples and consistent results with RT-PCR/PCR were produced in 105 samples. The agreement rate was 92%.
By using a viral nucleic acid multiplication method with high sensitivity, physicians can manage ALRIs with prolonged fever by only administering supportive care instead of switching to another type of antibiotics. This diagnostic method is being adopted for more accurate diagnosis, although the cost-effectiveness has still not been established. However, inappropriately prescribing antibiotics in viral respiratory infections would be resolved only by rigorous diagnosis [4]. In the present study, amoxicillin+clavulanate was the most frequently prescribed treatment for pneumonia. In case of adenovirus or influenza viral infections, prolonged fever was a striking feature, but we could wait without adding other antibiotics based on viral laboratory data and the coherent features of chest radiographs.
Questions remain as to whether multiplex PCR/RT-PCR methods are exclusively suitable for a respiratory surveillance network. Other methods, such as immunochromatography, direct fluorescent antibody (DFA) staining, or enzyme immunoassays, are producing compatible results to molecular studies in RSV, influenza virus, PIV (93–99%), and hMPV (85–95%) [3, 5–7]. If it is suitable, a commercial rapid kit could be used first as mass screening and in negative samples, and the adoption of a reflex PCR method could be considered. And the rapid culture kit also enables the physicians to make a timely diagnosis of conventional viral infections. Adenovirus and influenza viruses were isolated well in both rapid shell viral cultures and RT-PCR/PCR methods. In our study, about 92% agreement was obtained between PCR and rapid shell viral culture. In another study of similar populations, 80% concordance was report [8]. However, recently identified viruses such as hBoV, hMPV, RV, or hCoV are known as hardly cultured viruses but well detected by multiplex PCR/RT-PCR methods. So, the mass screening of respiratory viruses (including unknown viruses) for severe respiratory infections would be more accurate and desirable with viral nucleic acid extractions from samples and subsequent identification with primers [8, 9]. When only one of each viral gene was targeted, true positive samples could be missed by this method [10]. In this study, two kinds of primers were included in the multiplex RT-PCR kits for hMPV, hCoV, and influenza virus A and B.
Hospital-based surveillance for severe respiratory infections have significant merits. The transmission of SARS in the Toronto outbreak was nosocomial in 80% of cases [9]. Rapid viral diagnosis performed in the same hospital would be helpful for preventing nosocomial infections in immunocompromised patients and for more targeted management. In addition to this, strict diagnosis can affect treatment policy directly. If the government supports the total costs of molecular viral diagnosis in tertiary hospitals for national surveillance, early standardized broad respiratory samplings would be possible, regardless of insurance situations or patients’ economic states. Thus, this hospital-based surveillance network is superior to any other sentinel surveillance system in the aspects of appropriate sampling time, elapse period of sampling and examination, and feedback effects from the viral study.
In conclusion, this pilot study showed that respiratory viral detection using multiplex RT-PCR/PCR was feasible, with great benefits and few limitations to establish a national hospital-based surveillance system for severe lower respiratory infections, and enabled clinicians to anticipate clinical courses and helped using the minimal administration of antibiotics. For governments, a rigorous viral study might be a viable solution in reducing the high resistance and the total medical costs of antibiotics for severe respiratory infections.
Acknowledgments
We would like to thank Dr. Dong-Won Kang who made the database program for this study, Dr. Dae Ryong Kang who provided helpful consultations for the study design, precious members of the Korean Society of Pediatric Infectious Disease who recommended their brilliant opinions, and all of the members of the International Health Care Center at Severance Hospital who supported us in this study. This study was supported by intramural grant from Korea Centers for Disease Control and Prevention.
Competing interests
None to be declared.
References
- 1.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger A, Drosten Ch, Doerr HW, Stürmer M, Preiser W. Severe acute respiratory syndrome (SARS)—paradigm of an emerging viral infection. J Clin Virol. 2004;29:13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Syrmis MW, Whiley DM, Thomas M, Mackay IM, Williamson J, Siebert DJ, Nissen MD, Sloots TP. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004;6:125–131. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low D. Reducing antibiotic use in influenza: challenges and rewards. Clin Microbiol Infect. 2008;14:298–306. doi: 10.1111/j.1469-0691.2007.01910.x. [DOI] [PubMed] [Google Scholar]
- 5.Kikuta H, Sakata C, Gamo R, Ishizaka A, Koga Y, Konno M, Ogasawara Y, Sawada H, Taguchi Y, Takahashi Y, Yasuda K, Ishiguro N, Hayashi A, Ishiko H, Kobayashi K. Comparison of a lateral-flow immunochromatography assay with real-time reverse transcription-PCR for detection of human metapneumovirus. J Clin Microbiol. 2008;46:928–932. doi: 10.1128/JCM.01888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruest A, Michaud S, Deslandes S, Frost EH. Comparison of the Directigen flu A+B test, the QuickVue influenza test, and clinical case definition to viral culture and reverse transcription-PCR for rapid diagnosis of influenza virus infection. J Clin Microbiol. 2003;41:3487–3493. doi: 10.1128/JCM.41.8.3487-3493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landry ML, Cohen S, Ferguson D. Real-time PCR compared to Binax NOW and cytospin-immunofluorescence for detection of influenza in hospitalized patients. J Clin Virol. 2008;43:148–151. doi: 10.1016/j.jcv.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roh KH, Kim J, Nam MH, Yoon S, Lee CK, Lee K, Yoo Y, Kim MJ, Cho Y. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci. 2008;38:41–46. [PubMed] [Google Scholar]
- 9.Muller MP, Richardson SE, McGeer A, Dresser L, Raboud J, Mazzulli T, Loeb M, Louie M, Canadian SARS Research Network Early diagnosis of SARS: lessons from the Toronto SARS outbreak. Eur J Clin Microbiol Infect Dis. 2006;25:230–237. doi: 10.1007/s10096-006-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry ML, Cohen S, Ferguson D. Prospective study of human metapneumovirus detection in clinical samples by use of light diagnostics direct immunofluorescence reagent and real-time PCR. J Clin Microbiol. 2008;46:1098–1100. doi: 10.1128/JCM.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


