Abstract
A case–control study was designed to investigate the role of different Cryptosporidium spp. in Swedish dairy herds with and without calf diarrhoeal problems. Faecal samples were collected from preweaned calves, young stock and cows. Cryptosporidium oocysts were detected by sodium chloride flotation and epifluorescence microscopy. Molecular diagnostics were used to identify Cryptosporidium species. Samples containing C. parvum were further analysed to determine subtypes. Calf faecal samples were also analysed for rotavirus, coronavirus and Escherichia coli F5+. Total protein was assessed in 1- to 8-day-old calves. A questionnaire was used to identify differences in management routines. Cryptosporidium infection was diagnosed in all herds, with equal prevalence in case and control herds in all three age groups. Cryptosporidium parvum, Cryptosporidium bovis, Cryptosporidium ryanae and Cryptosporidium andersoni were all identified, as were rotavirus, coronavirus and E. coli F5+. C. ryanae and C. andersoni were only detected in non-diarrhoeal samples, whereas the other pathogens were detected in both diarrhoeal and non-diarrhoeal samples. Diarrhoea was more common in case herd calves. Disinfection of single pens was more common in case herds and several other management routines seemed to differ although results were not significant.
Keywords: Bovine Viral Diarrhoea Virus, Cryptosporidium Oocyst, Cryptosporidium Infection, Cryptosporidium Species, Young Stock
Introduction
Diarrhoea is a common problem in young calves and a cause of impaired animal welfare. Farmers may suffer large economic losses due to veterinary treatments, mortality and reduced growth rates. Calf diarrhoea has a multifactorial aetiology, and Cryptosporidium parvum is a protozoan parasite that is frequently associated with the disease (de la Fuente et al. 1999; Joachim et al. 2003; Snodgrass et al. 1986). The parasite is resistant in the environment (Fayer 2008), creating high infection pressures in some affected herds. Other common diarrhoeal pathogens are rotavirus, coronavirus and enterotoxigenic Escherichia coli F5+ (Barrington et al. 2002; de la Fuente et al. 1999; Foster and Smith 2009). E. coli F5+ causes diarrhoea in calves younger than 1 week, whereas the other three pathogens mainly cause diarrhoea in 1- to 3-week-old calves. Individual calf factors such as failure of passive transfer of colostral antibodies, and management associated factors such as personnel and stable hygiene, routines at calving, milk feeding regimens and crowding, affect the course of disease (Barrington et al. 2002).
A limitation with previously performed studies on the association of diarrhoea and cryptosporidiosis in calves is that all oocysts approximately 5 μm in diameter until recently were recognised as C. parvum. However, molecular diagnostic methods have shown that three morphologically similar species, C. parvum, Cryptosporidium bovis and Cryptosporidium ryanae, can be found in cattle. Several studies applying DNA analysis have identified C. parvum in 88.0–95.5% of preweaned calf diarrhoeal samples, with C. bovis and C. ryanae detected in 0–5.9% and 1.6–5.9% of samples, respectively (Plutzer and Karanis 2007; Soba and Logar 2008; Thompson et al. 2007). In contrast to C. parvum, C. bovis, C. ryanae and a fourth larger species, Cryptosporidium andersoni, are considered to be associated with subclinical infection in cattle (Fayer et al. 2007; Langkjaer et al. 2006). We have shown that Cryptosporidium parasites are ubiquitous in Swedish dairy herds but we found no association between infection and diarrhoea in individual calves (Silverlås et al. 2009). With the help of DNA analysis, we identified C. bovis as the most prevalent species (74%) in preweaned calves, and C. parvum, C. bovis and C. ryanae were all detected in diarrhoeal calves (Silverlås et al. 2010). It is thus not correct to conclude that diarrhoea is associated with C. parvum solely based on the presence of C. parvum-like oocysts. The improved diagnostic methods call for to further examine the relationship between Cryptosporidium infection and diarrhoea. It is also important to identify additional factors affecting the course of infection because a key component for improved calf health in herds with Cryptosporidium-associated diarrhoea is to decrease infection pressure by improved management.
The objective of this herd level case–control study was to investigate the role of Cryptosporidium infection, with the added value of information on the different Cryptosporidium spp. in Swedish dairy herds facing a problem with calf diarrhoea. To get a more complete picture of the diarrhoeal problems, other pathogens and animal and management factors were also investigated.
Materials and methods
Study design
Ten case herds and 10 control herds with ≥50 milking cows/year were included. Case herds were defined as herds that had current problems with diarrhoea in calves. Each case herd was matched to a control herd (without calf health problems) with approximately the same number of cows and situated in the same veterinary district. Possible case and control herds were identified by herd health veterinarians, who had followed these herds over several years and thus had a long-time insight into the calf health of each herd.
A questionnaire was designed to get information about herd structure and management factors believed to influence prevalence of Cryptosporidium-infected and diseased animals. Questions were grouped as follows: general herd information, routines at calving, calf management, disease in calves, weaned calf and young stock management. The questionnaire is available in Swedish from the first author upon request.
Sampling procedure and data collection
The study was conducted during the indoor seasons (mid October to end of March) 2006–2007 (n = 3 pairs) and 2007–2008 (n = 7 pairs), and each herd was visited once. Samplings were done by herd health veterinarians or by author CS. Matched case–control herds were sampled by the same veterinarian within a week from each other. A total of eight veterinarians participated. Detailed instructions about sampling procedures and data collection were sent to participating veterinarians to minimise personnel bias. Rectal faecal samples were collected from 10 calves (≤2 months old), 10 young stock (4–12 months old) and five periparturient cows (1 week before expected partus to 2 weeks post partum). Any additional calves with diarrhoea were also sampled. Eight herds had insufficient number of calves, young stock or periparturient cows at sampling, in which case all calves or young stock in the preset age interval and five cows as close to the preset interval as possible were sampled. Jugular vein blood was collected in serum tubes from a maximum of five 1- to 8-day-old calves in each herd. For sampled calves, a number of parameters, e.g. age, the consistency of faeces, general condition, pen environment and any medical treatment (drug, disease and dates of treatment) were recorded. For sampled young stock and cows, consistency of faeces and age were recorded. For cows the number of days in relation to calving was also recorded. The visiting veterinarian completed the questionnaires through interviews with the farmers.
Detection of Cryptosporidium oocysts and DNA analysis of Cryptosporidium positive samples
All faecal samples were processed by a saturated sodium chloride flotation method and analysed for Cryptosporidium oocysts by epifluorescence microscopy as described previously (Silverlås et al. 2009). The detection limit of this method is 50–100 oocysts per gram faeces (OPG) (Andersson 2004). Concentrated samples were stored at 4–8°C.
The aim was to analyse two samples positive for C. parvum-like oocysts (∼5 μm Ø) from each age group and herd. If more than two positive samples were present, random selection was done. All samples with the larger C. andersoni oocysts (∼5 × 7.5 μm) were analysed. DNA was extracted using a combined freeze-thawing and QIAamp DNA stool mini kit (QIAGEN) protocol. A nested PCR protocol for amplification of a ∼800-bp segment of the 18S rRNA gene and sequencing of PCR products was used for species determination. Samples containing C. parvum were subtyped through amplification of a ∼800-bp segment of the 60-kDa glycoprotein (GP60) gene using a nested PCR protocol and sequencing. All methods have previously been described (Silverlås et al. 2010).
Detection of other pathogens and control of colostral uptake
All calf faecal samples were investigated for presence of rota- and coronavirus, whereas presence of E. coli F5+ was investigated in samples from calves ≤14 days. Faeces for viral and bacteriological analyses were frozen at arrival to the laboratory, and thawed and analysed at the end of each sampling season. Total protein (TP) in blood was estimated the day samples arrived at the laboratory.
Rotavirus and coronavirus were detected by indirect antigen ELISAs. E. coli F5+ was detected by cultivation on blood agar and agglutination test of a few, randomly selected colonies. These methods are used in the routine diagnostics at the Swedish National Veterinary Institute (SVA).
Blood samples were centrifuged at 1500×g for 10 min at room temperature, and TP was assessed by refractometry of the serum. Calves with ≥55 g/L TP were considered to have sufficient colostral antibody uptake (Radostits 2000).
Statistical analysis
Statistical analysis was done using Stata 10 (© 1984–2008, StataCorp, College Station, TX, USA). An animal was considered Cryptosporidium positive if at least one C. parvum-like oocyst or C. andersoni oocyst was detected in the microscope. Statistical calculations were done at herd and calf level, using χ 2 test, Fisher’s exact test or the Mann–Whitney test. Epidemiological modelling was done at calf level to determine factors associated with being a case herd calf, using logistic regression with robust standard errors to adjust for clustering of observations within herds. Variables were first screened in univariable logistic regression models, and variables with p ≤ 0.25 were considered for further analyses. Collinearity between variables was investigated using Spearman rank correlations. Multivariable modelling was done by forward selection, using p ≤ 0.05 as a retaining criterion. Confounding was assessed as each variable was entered or deleted from the model. If the odds ratio (OR) of any variable category changed >25%, the confounding variable was retained in the model. When a final main effects model had been achieved, previously deleted variables were re-entered and evaluated again, and then two-way interactions were investigated. The final model was diagnosed using the Pearson goodness-of-fit test and the receiver operating characteristic curve, and plotting deviance and Pearson residuals against the linear prediction of residuals. Any detected outliers were investigated further.
Results
Eight of the case herds had a long-time history of diarrhoeal problems (at least since the preceding indoor season) and two herds had a sudden outbreak of diarrhoea and mortality. In total, 483 faecal samples from 100 cows, 187 young stock and 196 calves, and 47 blood samples were collected. Median age of all sampled calves was 25.5 days (n = 96) in control herds and 24 days (n = 114, range 2–62 days) in case herds (p = 0.68, Mann–Whitney test). Median age of sampled diarrhoeal calves was 20 days (n = 9, range 2–62 days) in control herds, and 29.5 days (n = 22, range 9–62 days) in case herds (p = 0.50, Mann–Whitney test).
Prevalence of Cryptosporidium oocysts
Cryptosporidium-positive animals were identified in all herds. Mono infection with C. parvum-like oocysts was identified in 208 animals and with C. andersoni oocysts in four animals. In addition, five animals had both C. andersoni and C. parvum-like oocysts, giving in total 217 Cryptosporidium-positive animals (44.9% prevalence). Overall mean prevalence in case herds was 45.7% and in control herds 41.7%. Group specific mean prevalences in case vs. control herds were: 61.6% vs. 64.6% in calves, 42.1% vs. 34.7% in young stock and 23% vs. 14% in cows. Neither overall mean prevalence nor group specific mean prevalences differed between case and control herds (p > 0.05, Mann–Whitney test). The prevalence of Cryptosporidium shedders in relation to age was similar in case and control herd calves, with a maximum prevalence in the third to fourth week of life in both groups (Fig. 1).
Fig. 1.
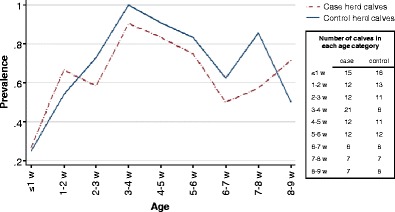
Prevalence of Cryptosporidium positive calves in case and control herds
Shedding rates of C. parvum-like oocysts were 100−2 × 108 OPG in calves (n = 129), 100−45,900 OPG in young stock (n = 70) and 100–650 OPG in cows (n = 14). Shedding rates of C. andersoni were 100–1,200 OPG in calves (n = 3), 100 OPG in young stock (n = 2) and 100–1.65 × 106 OPG in cows (n = 4).
Cryptosporidium species and C. parvum subtypes
Of 94 samples subjected to DNA analysis, 75 (80%) were successfully amplified at the 18S rRNA locus. Of these, 59 were C. bovis, eight were C. parvum, six were C. ryanae and two were C. andersoni.
C. bovis was identified in all herds, whereas C. parvum was identified in one control herd and four case herds, C. ryanae in two control herds and four case herds and C. andersoni in two case herds. Species distribution did not differ significantly between case and control herds (p > 0.05, Fisher’s exact test). C. bovis was identified in all age groups, C. ryanae in calves and young stock, C. parvum in calves and C. andersoni infection was only confirmed in cows (Table 1). Data on species distribution in individual animals from case and control herds are given in Table 1.
Table 1.
Distribution of detected pathogens in calves, young stock and cows
| Group | No diarrhoea | Diarrhoea | Totala | |||
|---|---|---|---|---|---|---|
| Pathogen | Case | Control | Case | Control | Case | Control |
| Calves (n = 196) | 82 | 83 | 22 | 9 | 104 | 92 |
| C. parvum | 4 | 2 | 2 | 0 | 6 | 2 |
| C. bovis | 7 | 17 | 5 | 1 | 12 | 18 |
| C. ryanae | 1 | 2 | 0 | 0 | 1 | 2 |
| C. andersoni | 0 | 3d | 0 | 0 | 0 | 3d |
| C. parvum-like spp.b | 40 | 38 | 10 | 2 | 50 | 38 |
| Rotavirus | 4 | 10 | 1 | 1 | 5 | 11 |
| Coronavirus | 0 | 4 | 0 | 2 | 0 | 6 |
| E. coli F5+ | 0 | 1 | 0 | 0 | 0 | 1 |
| Young stock (n = 187)c | 92 | 90 | 1 | 4 | 93 | 94 |
| C. parvum | 0 | 0 | 0 | 0 | 0 | 0 |
| C. bovis | 10 | 10 | 0 | 1 | 10 | 11 |
| C. ryanae | 2 | 1 | 0 | 0 | 2 | 1 |
| C. andersoni | 0 | 2d | 0 | 0 | 0 | 2d |
| C. parvum-like spp.b | 26 | 20 | 20 | |||
| Cows (n = 100)c | 50 | 50 | 0 | 0 | 50 | 50 |
| C. parvum | 0 | 0 | 0 | 0 | 0 | 0 |
| C. bovis | 5e | 3 | 0 | 0 | 5e | 3 |
| C. ryanae | 0 | 0 | 0 | 0 | 0 | 0 |
| C. andersoni | 2 (+2de) | 0 | 0 | 0 | 2 (+2de) | 0 |
| C. parvum-like spp.b | 3 | 4 | 0 | 0 | 3 | 4 |
a217 Cryptosporidium-positive animals, but the total number of Cryptosporidium-positive samples is 223 because of five samples with both C. parvum-like species and C. andersoni at microscopy and one sample positive for C. andersoni at microscopy, but for C. bovis, at DNA analysis, are counted twice
bSamples with C. parvum-like oocysts of unknown species (not used for DNA analysis or DNA analysis failed)
cSamples were not analysed for presence of rotavirus, coronavirus or E. coli F5+
dMicroscope analysis, could not be confirmed by DNA analysis
eOne sample positive for C. bovis at DNA analysis, but only a few C. andersoni oocysts detected by microscopy
All eight C. parvum samples were successfully sequenced at the GP60 locus. They belonged to subtypes IIaA16G1R1 (n = 5), IIaA17G1R1 (n = 2) and IIaA21G1R1 (n = 1). Subtype IIaA17G1R1 was the only one detected in diarrhoeal calves.
Rotavirus, coronavirus and E. coli F5+
Data on distribution of rotavirus, coronavirus and E. coli F5+ in calves from case and control herds are given in Table 1. Mono infection with rotavirus or coronavirus was present in four and two calves, respectively. Mixed infections were present in 16 calves as follows: rotavirus and C. parvum (n = 3), rotavirus and undetermined C. parvum-like spp. (n = 7), rotavirus and E. coli F5+ (n = 1), coronavirus and C. bovis (n = 1) and coronavirus and undetermined C. parvum-like spp. (n = 3).
Presence of diarrhoea and pathogens identified in diarrhoeic samples
Diarrhoea was present in 22 calves from nine of 10 case herds (herd prevalence 0–40%, median 20%) and nine calves from six of 10 control herds (herd prevalence 0–25%, median 10.6%) at sampling. A significantly higher proportion of case herd calves had diarrhoea (p = 0.03, χ 2), and herd prevalence differences approached statistical significance (p = 0.08, Mann–Whitney test). The case herd without diarrhoeal calves was one of the herds that suffered from an acute outbreak. The matched control herd also had only healthy calves at sampling. Pathogens detected in diarrhoeic calves were: C. parvum, C. bovis, undetermined C. parvum-like spp., rotavirus and coronavirus (Table 1). One of the diarrhoeic coronavirus-positive calves had a few C. parvum-like oocysts of undetermined species, but otherwise no co-infections were detected in diarrhoeic calves. In eight diarrhoeal samples, none of the investigated pathogens were detected.
Shedding rates for different Cryptosporidium species in diarrhoeic vs. non-diarrhoeic calves were 12,750−3 × 106 OPG vs. 2,800−2 × 108 OPG (C. parvum), 9,250−2 × 106 OPG vs. 1,200−1.65 × 106 OPG (C. bovis) and 100−3.5 × 106 OPG vs. 100−6.65 × 106 OPG (undetermined C. parvum-like spp.). No significant differences were found. In comparison, shedding rates in the non-diarrhoeic calves with C. ryanae were 3,650–500,000 OPG.
Management routines, total protein and mortality
Disinfection of single pens between calves was more common in case herds (p = 0.02, Fisher’s exact test). Used disinfectants were Virkon (Viroderm; n = 3), Stalosan F (Svenska Lantmännen, containing phosphates, kaolin, iron and copper compounds; n = 3), Stalosan F and Virkon (n = 1) and Stalosan F and powdered hydrated lime (n = 1). Several other differences in case and control herd management were indicated although significant differences were not detected or statistical tests could not be performed due to zero observations in some categories. These variables are presented in Table 2. All control herds fed colostrum by bottle, whereas routines varied in case herds. Two case herd and two control herd farmers routinely checked colostral quality. Total protein ranged from 42–71 g/L (median 55 g/L) in 25 case herd calves and from 44–66 g/L (median 57 g/L) in 22 control herd calves (p = 0.18, Mann–Whitney test). All farmers stated that they had storage of frozen colostrum.
Table 2.
General herd information, routines at calving and routines for care of preweaned calves
| Variable | Herds | P a | |
|---|---|---|---|
| Case | Control | ||
| General information | |||
| No. of cows/year | 42–202.6 | 50–165 | 0.36 |
| No. of calves at sampling | 13–45 | 8–35 | 0.17 |
| Average milk yield/cow and year (kg ECM) | 7,337–11,500 | 8,554–12,200 | 0.50 |
| Stall | |||
| Tiestall | 3 | 2 | |
| Cubicle housing | 5 | 8 | |
| Tie and cubicle | 2 | 0 | |
| Calving routines | |||
| Calving pen | |||
| Individual pen | 4 | 9 | |
| Group pen | 2 | 1 | |
| Tiestall | 4 | 0 | |
| Bedding in calving pen | |||
| Straw | 4 | 8 | |
| Wood shavings | 3 | 2 | |
| Straw + wood shavings | 3 | 0 | |
| Watches calvings | |||
| Around the clock | 7 | 4 | |
| No or during daytime | 3 | 6 | 0.37 c |
| Routines for first colostral intake | |||
| Calf manages on its own | 2 | 0 | |
| Gives bottle if needed | 5 | 0 | |
| Always give in bottle | 3 | 10 | |
| Amount of colostrum given at first meal | |||
| None or not specified | 6 | 1 | |
| <2 L | 0 | 3 | |
| ≥2 L | 4 | 6 | |
| Management of preweaned calves | |||
| Caretaker manages healthy calves on the same day as | |||
| Sick calves | 2 | 0 | |
| Sick older | 1 | 1 | |
| Sick calves and older | 7 | 9 | |
| Placing of single pens compared to group pens | |||
| Separate | 3 | 6 | |
| Close | 7 | 4 | 0.37 c |
| Cleaning of single pens | |||
| Several times/calf | 1 | 3 | |
| Between calves | 9 | 7 | 0.58 c |
| Disinfects single pens between calves | |||
| No | 3 | 9 | |
| Yes | 7 | 1 | 0.02 c |
| Age at transfer to group pens | |||
| <2 weeks of age | 4 | 4 | |
| ≥2 weeks of age | 5 | 3 | |
| No group pens | 1 | 3 | 0.64 c |
| Disease in calves | |||
| Dead of last 20 born alive | |||
| 0 | 4 | 9 | |
| 1 | 3 | 0 | |
| 2 | 2 | 0 | |
| 3 | 1 | 1 | |
| Most common type of diarrhoea | |||
| Pasty | 1 | 5 | |
| Watery | 6 | 4 | |
| Varies considerably | 3 | 1 | 0.21 c |
| Has treated sampled calves against | |||
| Nothing | 5 | 9 | |
| Acute diarrhoea | 1 | 0 | |
| Prophylaxis (diarrhoea) | 2 | 0 | |
| Pneumonia | 2 | 1 | |
aFor variables lacking a p value, calculations were not done because of zero observations in one or more cells
bMann–Whitney test
cTwo-sided Fisher’s exact test
Neonatal mortality was registered as number of deaths before weaning in the last 20 calves born alive. Calves had died in six case herds and in one control herd (Table 2). Mortality was mostly due to diarrhoea, but respiratory disease, trauma and unknown causes were also registered. No calves had been treated prophylactically or therapeutically against cryptosporidiosis prior to sampling.
Model of factors associated with being a case herd calf
Seven variables had p ≤ 0.25 in univariable modelling (Table 3). The multivariable regression model contained three significant variables and one confounder (Table 3). ‘C. bovis status’ gave reduced odds of belonging to a case herd, but calves with unknown C. bovis status (undetermined C. parvum-like spp.) did not differ from calves negative for C. bovis (i.e. Cryptosporidium negative or infected with C. parvum or C. ryanae). The variable ‘C. parvum identified in herd’ did not produce a significant odds ratio, but acted as a confounder for both ‘disinfection of single pens’ and ‘most common type of diarrhoea’.
Table 3.
Variables with p ≤ 0.25 in univariable logistic regression models and final multivariable logistic regression model of factors associated with being from a case or control herd in 210 calves, using robust standard errors to adjust for 20 herd clusters
| Variable | Univariable model | Multivariable modela | ||||
|---|---|---|---|---|---|---|
| Observations | P | Odds ratio | P | 95% confidence interval | ||
| Case | Control | |||||
| Calf level | ||||||
| Box type | 0.154 | – | – | – | ||
| Single pen or hut | 51 | 64 | ||||
| Group box | 55 | 28 | ||||
| C. bovis status | 0.047 | 0.004b | ||||
| Negative | 43 | 35 | 1 | |||
| Positive | 10 | 17 | 0.48 | 0.008 | 0.28; 0.82 | |
| Unknown | 61 | 44 | 0.81 | 0.633 | 0.34; 1.93 | |
| Herd level | ||||||
| C. parvum identified in herd | 0.175 | 0.489b | ||||
| No | 69 | 86 | 1 | |||
| Yes | 45 | 10 | 2.9 | 0.489 | 0.15; 55.80 | |
| Disinfects single pens between calves | 0.035b | |||||
| No | 34 | 88 | 0.012 | 1 | ||
| Yes | 80 | 8 | 35.6 | 0.035 | 1.29; 978.84 | |
| Most common consistency of diarrhoea | 0.240 | 0.015b | ||||
| Pasty | 11 | 44 | 1 | |||
| Watery | 67 | 41 | 4.1 | 0.209 | 0.45; 36.7 | |
| Varies considerably | 36 | 11 | 47.3 | 0.012 | 2.35; 951.44 | |
| Placing of single pens relative to group boxes | 0.192 | – | – | – | ||
| Separate | 33 | 57 | ||||
| In proximity to group boxes | 81 | 39 | ||||
| Surveillance of calvings | 0.149 | – | – | – | ||
| Around the clock | 35 | 62 | ||||
| None or daytime | 79 | 34 | ||||
aWald χ2 = 43.40, p < 0.001, pseudo R2 = 0.4838; Pearson goodness-of-fit test, χ2 25.7, p = 0.016
bOverall p value of variable
Odds of being a case herd calf were higher if the type of diarrhoea varied considerably, compared to if pasty diarrhoea was most common in a herd. There was also a tendency of higher odds in calves from herds where watery diarrhoea was most common. When a farmer reported to routinely disinfect single pens between calves, the odds of being a case herd calf were higher than if disinfection was not a routine. No significant interactions were detected. The model had a rather poor fit (Table 3), but the area under the receiver operated characteristics curve was 0.91 (maximum 1) and no residual outliers were detected.
Discussion
Presence of Cryptosporidium and other pathogens
The complexity of Cryptosporidium infection and its association to clinical symptoms is reflected in that we found Cryptosporidium-infected calves in all investigated herds and similar shedding rates of C. parvum, C. bovis and undetermined C. parvum-like spp. in diarrhoeic and non-diarrhoeic calves, which shows that massive C. parvum infection is not sufficient to produce diarrhoea. Shedding rates approximately follow bell shaped curves over time, and shedding intensity and diarrhoeal intensity seem to peak around the same time (Jarvie et al. 2005; Joachim et al. 2003; Lallemond et al. 2006; Lefay et al. 2001; Villacorta et al. 1991). For calves with high C. parvum shedding rates, it is thus likely that the parasite is associated with diarrhoea, and the identification of high numbers of C. bovis oocysts in diarrhoeic samples indicates that this species can cause diarrhoea in calves. C. bovis has been associated with subclinical infection (Santín et al. 2004), but massive intestinal invasion with subsequent diarrhoea should be possible if enough other factors, such as lack of passive immunity, are present, even if this species would lack virulence factors similar to those present in C. parvum.
The interpretation of diarrhoeal samples containing a few oocysts is not as straightforward, especially when such a sensitive diagnostic method as ours is used and only one sample per calf is collected. Because intestinal damage is caused by the intracellular parasite stages, diarrhoea might be present already when the first few oocysts are shed because these do not reflect the intensity of intestinal invasion. Moreover, calves with failure of passive transfer of colostral antibodies have poor health to start with, and might develop diarrhoea more easily and at a lower degree of intestinal invasion than calves with sufficient passive transfer. On the other hand, if the few oocysts reflect low-grade infection, the real cause for diarrhoea could be an unidentified pathogen. Repeated samplings are crucial to confirm or reject the association between infection and diarrhoea in these calves, but that could not be done in this study.
We have earlier identified the C. parvum subtypes IIaA16G1R1 and IIaA21G1R1 found here in Swedish cattle (Silverlås et al. 2010), whereas subtype IIaA17G1R1 was first identified in this study. We found subtype IIaA16G1R1 in one non-diarrhoeic calf in the preceding study (Silverlås et al. 2010) as well as in five non-diarrhoeic calves from this study, but this subtype and IIaA17G1R1 have both previously been identified in diarrhoeic calf samples (Plutzer and Karanis 2007; Soba and Logar 2008; Thompson et al. 2007). We were first to report subtype IIaA21G1R1 in two calves (Silverlås et al. 2010), and this subtype has now been identified in three non-diarrhoeic calves in total. The GP60 locus used to identify these C. parvum subtypes encodes a glycoprotein situated in the apical region of invasive stages of the parasite. GP60 subtypes are also identified in the human species C. hominis and have been associated with varying pathogenicity in HIV patients (Cama et al. 2007). Due to the low number of subtyped samples in this study, conclusions about pathogenicity or association with diarrhoeal problems cannot be made.
This study was designed at herd level, and diarrhoeic calf samples were not targeted for DNA analysis. Still 40% of diarrhoeic Cryptosporidium-positive calf samples were sequenced, compared to 28% of non-diarrhoeic samples. Although only two Cryptosporidium-positive calf samples per herd were subjected to DNA analysis, these represented a random sample and should not bias the species distribution.
At least one of the investigated pathogens were detected in most diarrhoeic samples but an equal or even higher percentage of the non-diarrhoeic calves also harboured them. In addition, only one of 16 calves with mixed infections had diarrhoea. About half of all diarrhoeic calves were older than 4 weeks. This indicates that other diarrhoeal causes such as Eimeria spp., or dietary imbalances could also be present in the herds of this study. However, diarrhoeal pathogens outside the “C. parvum diarrhoeal window” were not targeted because the main focus was to investigate the role of different Cryptosporidium species in herds with diarrhoeal problems. The reason for including calves older than 4 weeks is that we have shown Cryptosporidium infection to be common in this age group (Silverlås et al. 2009). Preweaned calves are often kept close together so these calves may act as an important infection source for the younger ones. Indeed, a high prevalence in old calves was shown in this study as well (Fig. 1). Young stock and cows were also included to get an impression of the overall Cryptosporidium infection pressure in the herds.
Even though rotavirus and coronavirus are considered major diarrhoeal pathogens in calves, they were uncommon also in case herds in the present study. We have no explanation for the results regarding rotavirus. We used the same ELISA, and the same person performed the analysis as in two Swedish studies that identified rotavirus in 24% and 44% of diarrhoeal samples, and in 8.9% and 3.7% of non-diarrhoeal samples (Björkman et al. 2003; de Verdier Klingenberg and Svensson 1998). The coronavirus results are not so surprising. Björkman et al. (2003) found a low prevalence (3%) of coronavirus-positive diarrhoeal samples, and in a recent Norwegian study (Gulliksen et al. 2009) no diarrhoeal samples were positive for coronavirus. In addition, our results are supported by Snodgrass et al. (1986) who found coronavirus to be equally common in healthy and diseased calves. The low prevalence of E. coli F5+-positive samples was in accordance with previous studies (Björkman et al. 2003; de Verdier Klingenberg and Svensson 1998). Other pathogens such as Salmonella spp. have been reported as common calf diarrhoeal pathogens (Barrington et al. 2002). In addition, bovine viral diarrhoea virus (BVDV) is recognised as an important calf health modulator worldwide, among other things causing immunosuppression and diarrhoea. As a result of successful control programmes, BVDV and Salmonella are uncommon in Swedish cattle, with 99.8% of herds being free from BVDV (professor Stefan Alenius, personal communication), and Salmonella only being identified in five to 13 herds per year (Anonymous 2007).
Although diarrhoea was more common in case herd calves, in total only 31 of 196 sampled calves had diarrhoea at sampling. This could be due to naturally fluctuating infection pressures. Even if diarrhoeal prevalences were quite low in case herds and pathogen distribution was similar in healthy and diarrhoeal calves from case and control herds, the identification of herds as cases or controls were performed by veterinarians with insight into the herds’ calf health status. We thus are confident that cases and controls were correctly classified. Ten case–controls is a small sample size, but a larger sample was not possible for logistic reasons.
Management routines
Several variables indicated differences between case and control herds (Table 2), and more case herd calves had diarrhoea compared to control herd calves. At a glance, individual calving pens, colostral feeding by bottle etc were more common in control herds, indicating better management routines and thus better biosecurity. Management routines in case herds also seemed to vary more than in control herds (Table 2). All case herd farmers applied at least one good management routine, but which one(s) differed. This indicates that different routines have to be addressed to improve biosecurity in the case herds.
Because of the differences in colostrum feeding strategies between case and control herds, we did not expect median TP values to be comparable. Both case and control herd calves had median TPs close to the acceptable value of 55 g/L (Radostits 2000), which shows that approximately half of all calves had insufficient colostral uptake. Colostral routines obviously needed improvement, but just as much in control herds as in case herds.
A Swedish study including 8,964 heifer calves noted a 3.1% mortality rate in the first 90 days from birth (Svensson et al. 2006). Accepting a 5% mortality rate (i.e. one of 20), three case herds and one control herd had elevated mortality rates (10–15%).
An interesting finding was that disinfection of single pens between calves was much more common in case herds. The same association has been shown for weaners (Maddox-Hyttel et al. 2006). This association could be seen as an effect—i.e. herds with diarrhoeal problems start to disinfect. However, because these herds still had problems, disinfection seemed either not to work or to be insufficient. Moreover, if disinfection routines were started a short time prior to sampling, the full effect might not have been reached at the time of sampling. We did not ask about how long these routines had been used, and cannot investigate this possibility further because of the high risk of recall bias. A potential explanation for disinfection not working is that farmers rely on the disinfectant to do the work of reducing infection pressure instead of cleaning thoroughly. Without proper cleaning, only the surface of the dirt layer is disinfected, pathogens can continue to thrive beneath, and calves are exposed again as soon as the top layer is disrupted. Cryptosporidia are resistant to most commercial disinfectants (Fayer 2008). Thus, if cryptosporidia constitute a large part of the problem even proper disinfection that reduces viral and bacterial pathogens would not be successful. Of the disinfectants used by farmers in this study, Virkon has not proven effective against Cryptosporidium oocysts (Fayer 2008, Table 1.9). Lime seems to reduce oocyst numbers (Graczyk et al. 2008; Zintl et al. 2010). These studies used quicklime (CaO) on sewage sludge (Graczyk et al. 2008) or hydrated lime (Ca[OH]2) in water solution and purified oocyst solutions incubated in used bedding material (Zintl et al. 2010). The effect of powdered hydrated lime on faecal oocysts has to our knowledge not been tested. Still, this might be at least partially effective in reducing oocyst load. We have not been able to identify references concerning the components of Stalosan F. Drying and heating are effective for decreasing oocyst viability (Fayer 2008, Table 1.7). Thus, proper cleaning (with water), followed by drying and use of powdered quicklime, or the use of deep litter bedding (provided that the surface is regularly overlaid with dry, clean bedding material) might be two effective ways of keeping Cryptosporidium infection pressure down.
Model of factors associated with being a case herd calf
Calves infected with C. bovis had lower odds of belonging to a case herd, indicating that C. bovis is more common in herds without diarrhoeal problems, and that C. parvum is really the species associated with diarrhoea. The fact that disinfection of single pens was identified as a risk factor could have several explanations that have already been discussed. The type of diarrhoea most common in preweaned calves could also be seen as an effect rather than a risk for belonging to a case herd. This variable was introduced in the model as an indicator of total infection pressure, hypothesising that a low total infection pressure would be more associated with pasty diarrhoea and that a high total infection pressure would be more associated with watery diarrhoea. Because some farmers stated that the type of diarrhoea varied considerably, this represented a third level of the variable. Indeed, although the level watery diarrhoea tended to have higher OR, this was the level that produced a significantly higher OR compared to pasty diarrhoea. A highly fluctuating infection pressure, due to e.g. long time between thorough cleaning of pens or fluctuations in calving intensity, could explain the significant association.
Concluding remark
Cryptosporidium infection was equally common in case and control herds, showing that these parasites can be abundant without causing diarrhoeal problems. Our results emphasise that management factors are important to whether diarrhoea will develop or not after infection with one or more of the investigated pathogens, including the pathogenic Cryptosporidium species C. parvum.
Acknowledgements
This study was supported financially by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, the Ivar and Elsa Sandberg Foundation, the Albert Hjärre foundation and Djurvännernas förening i Stockholm. We acknowledge Katarina Näslund, Helena Reineck and Anna Rydzik for skillful work in the laboratory. We would also like to thank veterinarians and farmers who participated in this study.
Ethical standards
The authors declare that the experiments comply with the current laws of the country in which they were performed. The study was approved by the Ethical Committee for Animal Experimentation in Uppsala, Sweden.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- Andersson S (2004) Kryptosporidieinfektion hos nötkreatur-utvärdering av en ny metod för påvisande av subklinisk infektion. Master of Veterinary Medicine thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden
- Anonymous (2007) Svensk zoonosrapport 2007. http://www.sva.se/upload/pdf/Tj%C3%A4nster%20och%20produkter/Trycksaker/SVA_zoonosrapport2007_webb.pdf. Accessed June 30 2009
- Barrington GM, Gay JM, Evermann JF. Biosecurity for neonatal gastrointestinal diseases. Vet Clin North Am Food Anim Pract. 2002;18:7–34. doi: 10.1016/S0749-0720(02)00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman C, Svensson C, Christensson B, de Verdier K. Cryptosporidium parvum and Giardia intestinalis in calf diarrhoea in Sweden. Acta Vet Scand. 2003;44:145–152. doi: 10.1186/1751-0147-44-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- de la Fuente R, Luzon M, Ruiz-Santa-Quiteria JA, Garcia A, Cid D, Orden JA, Garcia S, Sanz R, Gomez-Bautista M. Cryptosporidium and concurrent infections with other major enterophatogens in 1 to 30-day-old diarrheic dairy calves in central Spain. Vet Parasitol. 1999;80:179–185. doi: 10.1016/S0304-4017(98)00218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Verdier KK, Svensson L. Group A rotavirus as a cause of neonatal calf enteritis in sweden. Acta Vet Scand. 1998;39:195–199. doi: 10.1186/BF03547792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer R. General biology. In: Fayer R, Xiao L, editors. Cryptosporidium and cryptosporidiosis. 2. Boca Raton: CRC; 2008. pp. 1–42. [Google Scholar]
- Fayer R, Santin M, Trout JM. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet Parasitol. 2007;145:260–266. doi: 10.1016/j.vetpar.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Foster DM, Smith GW. Pathophysiology of diarrhea in calves. Vet Clin North Am Food Anim Pract. 2009;25:13–36. doi: 10.1016/j.cvfa.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk TK, Kacprzak M, Neczaj E, Tamang L, Graczyk H, Lucy FE, Girouard AS. Occurrence of Cryptosporidium and Giardia in sewage sludge and solid waste landfill leachate and quantitative comparative analysis of sanitization treatments on pathogen inactivation. Environ Res. 2008;106:27–33. doi: 10.1016/j.envres.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Gulliksen SM, Jor E, Lie KI, Hamnes IS, Loken T, Akerstedt J, Osteras O. Enteropathogens and risk factors for diarrhea in Norwegian dairy calves. J Dairy Sci. 2009;92:5057–5066. doi: 10.3168/jds.2009-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvie BD, Trotz-Williams LA, McKnight DR, Leslie KE, Wallace MM, Todd CG, Sharpe PH, Peregrine AS. Effect of halofuginone lactate on the occurence of Cryptosporidium parvum and growth of neonatal dairy calves. J Dairy Sci. 2005;88:1801–1806. doi: 10.3168/jds.S0022-0302(05)72854-X. [DOI] [PubMed] [Google Scholar]
- Joachim A, Krull T, Schwarzkopf J, Daugschies A. Prevalence and control of bovine cryptosporidiosis in German dairy herds. Vet Parasitol. 2003;112:277–288. doi: 10.1016/S0304-4017(03)00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemond M, Villeneuve A, Belda J, Dubreuil P. Field study of the efficacy of halofuginone and decoquinate in the treatment of cryptosporidiosis in veal calves. Vet Rec. 2006;159:672–677. doi: 10.1136/vr.159.20.672. [DOI] [PubMed] [Google Scholar]
- Langkjaer RB, Vigre H, Enemark HL, Maddox-Hyttel C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 2006;89:1–12. doi: 10.1017/S0031182006001533. [DOI] [PubMed] [Google Scholar]
- Lefay D, Naciri M, Poirier P, Chermette R. Efficacy of halofuginone lactate in the prevention of cryptosporidiosis in suckling calves. Vet Rec. 2001;148:108–112. doi: 10.1136/vr.148.4.108. [DOI] [PubMed] [Google Scholar]
- Maddox-Hyttel C, Langkjaer RB, Enemark HL, Vigre H. Cryptosporidium and Giardia in different age groups if Danish cattle and pigs—occurrence and management associated risk factors. Vet Parasitol. 2006;141:48–59. doi: 10.1016/j.vetpar.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Plutzer J, Karanis P. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet Parasitol. 2007;146:357–362. doi: 10.1016/j.vetpar.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Radostits OM (2000) The principles of control of infectious diseases of calves under 30 days of age. In: the World Buiatrics Association (ed) Proceedings of the XXI World Buiatrics Congress, Punta del Este, Uruguay, 4–8 Dec 2000, pp 1521–1556
- Santín M, Trout J, Xiao L, Zhou L, Greiner E, Fayer R. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol. 2004;122:103–117. doi: 10.1016/j.vetpar.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Silverlås C, Emanuelson U, de Verdier K, Björkman C. Prevalence and associated management factors of Cryptosporidium shedding in 50 Swedish dairy herds. Prev Vet Med. 2009;90:242–253. doi: 10.1016/j.prevetmed.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Silverlås C, Näslund K, Björkman C, Mattsson JG. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol. 2010;169:289–295. doi: 10.1016/j.vetpar.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Snodgrass DR, Terzolo HR, Sherwood D, Campbell I, Menzies JD, Synge BA. Aetiology of diarrhoea in young calves. Vet Rec. 1986;119:31–34. doi: 10.1136/vr.119.2.31. [DOI] [PubMed] [Google Scholar]
- Soba B, Logar J. Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology. 2008;135:1263–1270. doi: 10.1017/S0031182008004800. [DOI] [PubMed] [Google Scholar]
- Svensson C, Linder A, Olsson SO. Mortality in Swedish dairy calves and replacement heifers. J Dairy Sci. 2006;89:4769–4777. doi: 10.3168/jds.S0022-0302(06)72526-7. [DOI] [PubMed] [Google Scholar]
- Thompson HP, Dooley JS, Kenny J, McCoy M, Lowery CJ, Moore JE, Xiao L. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol Res. 2007;100:619–624. doi: 10.1007/s00436-006-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villacorta I, Peeters JE, Vanopdenbosch E, Ares-Mazás E, Theys H. Efficacy of halofuginone lactate against Cryptosporidium parvum in calves. Antimicrob Agents Chemother. 1991;35:283–287. doi: 10.1128/aac.35.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintl A, Keogh B, Ezzaty-Mirhashemi M, De Waal T, Scholz D, Mulcahy G. Survival of Cryptosporidium parvum oocysts in the presence of hydrated lime. Vet Rec. 2010;166:297–300. doi: 10.1136/vr.c1157. [DOI] [PubMed] [Google Scholar]


