Abstract
The recent emergence and re-emergence of alphaviruses, in particular the chikungunya virus (CHIKV), in numerous countries has invoked a worldwide threat to human health, while simultaneously generating an economic burden on affected countries. There are currently no vaccines or effective drugs available for the treatment of the CHIKV, and with few lead compounds reported, the vital medicinal chemistry is significantly more challenging. This study reports on the discovery of potential inhibitors for the nsP3 macro domain of CHIKV using molecular docking, virtual screening, and molecular dynamics simulations, as well as work done to evaluate and confirm the active site of nsP3. Virtual screening was carried out based on blind docking as well as focused docking, using the database of 1541 compounds from NCI Diversity Set II, to identify hit compounds for nsP3. The top hit compounds were further subjected to molecular dynamic simulations, yielding a greater understanding of the dynamic behavior of nsP3 and its complexes with various ligands, concurrently confirming the outcomes of docking, and establishing in silico lead compounds which target the CHIKV nsP3 enzyme.
Figure.
Virtual screening identifies novel inhibitors targeting the nsP3 macro domain of chikungunya virus
Electronic supplementary material
The online version of this article (doi:10.1007/s00894-014-2216-6) contains supplementary material, which is available to authorized users.
Keywords: Chikungunya virus, Molecular docking, Virtual screening, Molecular dynamics simulations, nsP3 macro domain
Introduction
Chikungunya virus (CHIKV) is an emerging mosquito-borne arthrogenic member of the Alphavirus genus from the family Togaviridae, which has caused widespread outbreaks of debilitating human disease in the last 5 years [1]. The resulting chikungunya fever (CHIKF) was first described in 1952 [2], and has been identified in nearly 40 countries. It was listed in 2008 as a US National Institute of Allergy and Infectious Diseases (NIAID) category C priority pathogen due to its high morbidity and mortality rates and thus major impact on health [3, 4].
After the initial mosquito bite, symptoms of chikungunya fever infection generally start in 4–7 days, and usually occur in two phases: an acute followed by a second persistent (chronic) stage that causes disabling polyarthritis [5]. Acute infection lasts 1–10 days and is characterized by a painful polyarthralgia, high fever, asthenia (weakness), headache, vomiting, rash, and myalgia [6]. The persistent chronic stage of CHIKF is characterized by polyarthralgia lasting from weeks to years [7]. Additional symptoms include neurological disorders such as encephalitis, myeloneuropathy, peripheral neuropathy, myelopathy, and myopathy [8].
The CHIKV genome consists of a single-stranded, positive-sense RNA genome with two open reading frames (ORFs) [9], and is approximately 11.8 kb in size. The ORF at the 5′ end encodes two polyproteins, the precursors of the nonstructural proteins. The second ORF at the 3′ end encodes the structural proteins, the capsid (C), envelope glycoproteins E1 and E2, and two small cleavage products (E3, 6 K). CHIKV starts its life cycle by entering the target host cells through pH-dependent endocytosis via a receptor-mediated interaction [10] in a manner analogous to that seen for other members of the Alphavirus genus.
The medicinal chemistry of CHIKV has recently been reviewed [11]. That review highlighted the extreme lack of available chemotherapeutics that show any inhibitory effects against the virus. In contrast, the emergence of numerous models and solved crystal structures of CHIKV proteins points to enormous possibilities for directed drug design. Notable among the available targets are the envelope proteins [12–15] and the nonstructural proteins of CHIKV, which play an important role in the formation of the transcription/replication complex of the virus [15, 16]. Among these, nsP3 is considered an attractive target for drug design because of its participation in the early stage of the transcription process of viral replication, even though the specific functions, roles, and activities of the nsP3 protein remain elusive [16, 17].
To date, there have been relatively few studies on the functions, roles, and activities of alphavirus nsP3 proteins [11, 18–20]. Studies based on the Sindbis virus reported that nsP3 phosphoprotein is an essential component of the viral replication and transcription process. Functional analysis of the effects of mutations of nsP3 on RNA synthesis demonstrated that the mutations can cause a loss of capacity for minus-strand synthesis or a failure to increase plus-strand synthesis. Strikingly, a change in G4303 implying an alteration from Gly to Ala68 and leading to a modification to the His-Ala-Val peptide was predicted to form part of the active site of the conserved nsP3 macro domain [19]. However, no effect of the mutations on the ADP-ribose binding site was found [17]. The nsP3 protein consists of two domains, the N-domain and the C-domain [16, 17]; the N-domain is highly conserved but the C-domain is not [17]. The C-domain is phosphorylated at up to 16 positions on serines and threonines [11, 17]. The role of this phosphorylation is still not clear, but deleting the residues involved in the phosphorylation process can decrease the level of RNA synthesis [11, 17, 21]. The N-domain, in which the region comprising the first 160 residues is called the X-domain or a macro domain, is commonly present in eukaryotic organisms, bacteria, archaea, and in many positive-strand RNA viruses such as hepatitis E, rubella, coronaviruses, and alphaviruses [16]. The alphavirus macro domain has a highly positively charged patch on the surface of the protein at the crevice of the ADP-ribose 1″-phosphate active site and its periphery [17]. The other side of the protein, far from the active site, possesses a negative charge. Thus, the nsP3 macro domain is considered to complex with ADP-ribose derivatives and RNA. It is also believed to control the metabolism of ADP-ribose 1″-phosphate and/or other ADP-ribose derivatives with regulatory functions in the cell [11, 22].
The crystal structure of the nsP3 macro domain of CHIKV was solved in 2010 [17]. The asymmetric unit of CHIKV includes four molecules. The macro domain consists of six-stranded β-sheets and four α-helices, and the positions of the α-helices are highly conserved. Also present is the ligand ADP-ribose, and the active site of the nsP3 macro domain is considered the binding site for this ADP-ribose ligand [16, 17]. This crevice is at the top of β-strands 2, 4, and 5 and is surrounded by two loops between β2-α1 and β5-α3. Since there is very little information available on the nsP3 macro domain and its inhibitor, the current study used computational approaches (including molecular docking and virtual screening) to discover potential compounds that inhibit nsP3 in CHIKV. Furthermore, in order to understand both the static structures and the dynamic information, molecular dynamics simulations were performed to gain a greater understanding of the behavior of nsP3 and how it changes upon the binding of small molecules. The results of the study were analyzed and compared with previous data to gain a deeper understanding of the atomic structure of the nsP3 macro domain and its complexes, as well as to confirm the interactions and results from docking.
Material and methods
Molecular docking and virtual screening
Molecular docking and virtual screening were conducted using AutoDock Vina (version 1.5.4) [23]. In the current study, the protein was treated as rigid, while the ligands were fully flexible. The protein was prepared by retrieving the 3D crystal structure of the nsP3 macro domain protein from the Protein Data Bank (PDB id: 3GPG), and this was used as the receptor for docking. The ligand ADP-ribose was extracted from the complex crystal structure (PDB id: 3GPO), whereas other ligands employed for virtual screening were downloaded from the website of the Diversity Set II chemical library of the National Cancer Institute (NCI). This library was selected as it contains diverse drug-like compounds, and potential hits can be ordered from the Open Chemical Repository Collection for activity testing. The protein was subsequently minimized by applying the CHARMM force field in the Accelrys Discovery Studio 2.0 software package [24]. The steepest-descent algorithm with 3,000 steps was used to relax the structure and remove steric overlaps. With a Cα RMSD of 0.59 Å between the minimized structure and the X-ray structure, the minimized structure was utilized for the docking process. Polar hydrogen atoms were added and the Kollman charges were defined with AutoDock Tools (version 1.5.4). The key docking parameters include the location of the docking site (center x, y, z) and the size of the grid box. The docking protocol was established by re-docking the ADP-ribose into nsP3 as seen in the co-crystal structure (3GPO). In the docking with ADP-ribose, box dimensions of 16 × 16 × 16 Å were used, centered at the crystallographically determined binding site. In virtual screening, the location and the size of the grid box were carefully investigated via blind docking (in which the box is sufficiently large to cover the whole protein) and focused docking (in which a smaller box was centered on the potential binding site of interest). Blind docking can reveal potential binding sites in nsP3, but it can also suffer convergence problems during sampling. The MetaPocket program was also used to predict potential binding sites of nsP3 other than the active site [25]. After docking, the results were analyzed and compared with available experimental data. The top ten compounds (“hits”) were identified according to their binding affinities using the default scoring function in Vina. The modes of interaction between the ligands and the protein were analyzed in Accelrys Discovery Studio 2.0. The drug-like properties of these compounds were subsequently evaluated using the Lipinski guidelines [26].
Molecular dynamics simulations of the docked complexes
In order to evaluate the stability of the docked complex and better characterize the ligand–nsP3 interactions, equilibrium molecular dynamics (MD) simulations were carried out with the NAMD package [27]. The protein atoms were treated with the CHARMM22 force field [28], and the corresponding parameters for the ligands were generated with AmberTools [29]. MD simulations were carried out for the apo state (PDB id: 3GPG) and the five ligand–nsP3 complexes. The systems were solvated in a cubic box of TIP3P water molecules and neutralized by sodium counterions to achieve the physiological ionic concentration of 0.15 M with NaCl. The total number of atoms (including protein, water, and counterions) in the CHIKV system was about 18,000, and the size of the water box was about 58 × 58 × 58 Å. All simulations were performed under periodic boundary conditions at a temperature of 298.15 K and a pressure of 1 atm. Temperature and pressure coupling were maintained with the Langevin algorithm. A particle mesh Ewald algorithm was used to calculate electrostatic interactions [30]. The cutoff distance for van der Waals interactions and the pair-list distance were 12 and 13.5 Å, respectively. The systems were minimized and then equilibrium simulations with weak harmonic restraints on the heavy atoms were performed for 3 ns. The production runs were continued for 20 ns. The trajectories were saved every 1 ps. The resulting trajectories were analyzed using the CHARMM [31] and VMD 1.9 programs [32].
Results
Validation of the docking protocol by re-docking with ADP-ribose
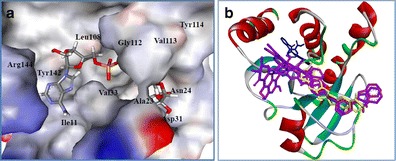
The docking protocol was evaluated by re-docking ADP-ribose into nsP3, and the outcome was compared with the available co-crystal structure (PDB id: 3GPO). The ligand conformations were ranked in terms of their predicted binding affinities using the default scoring function in Vina (see Table S1 of the “Electronic supplementary material,” ESM). The best-docked pose had a binding affinity of −10.2 kcal mol−1. The hydrogen bonds formed between ADP-ribose and nsP3 are shown in Fig. 1. It is evident that there were interactions with 11 residues in the active site of nsP3: Ile11, Ala23, Asn24, Asp31, Val33, Leu108, Gly112, Val113, Tyr114, Tyr142, and Arg144. Most of the hydrogen-bond donors came from the protein residues, with the corresponding acceptors deriving from the ADP-ribose. In addition, the diphosphate component of ADP-ribose showed the strongest interaction of any part of this ligand.
Fig. 1a–b.
Re-docking ADP-ribose into the active site of nsP3. a The best docking pose of ligand ADP-ribose is represented as a stick model (colored by atom type), while the protein nsP3 is shown in the solvent surface (colored by interpolated charge, with a probe radius of 1.4 Å). b The interactions of this pose and nsP3 residues showing hydrogen-bonding interactions at the binding site of nsP3
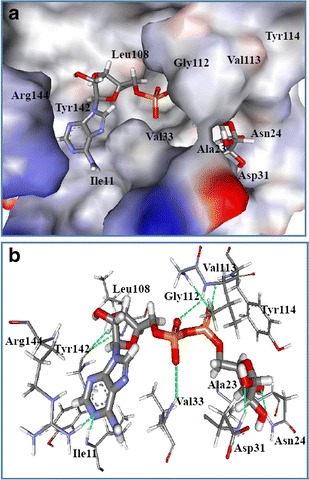
Moreover, the accuracy and reliability of docking were evaluated by superimposing the docked structure on the co-crystal structure. The heavy-atom root mean square deviation (RMSD) was 0.6 Å, smaller than the 2.0 Å often used as a criterion in correct bound structure prediction [33] (Table S1 of the ESM and Fig. 2), indicating that the molecular docking reproduced the binding mode in the co-crystal structure. Comparison of the interactions indicated by the docking results with published data (Table 1) confirmed there was good agreement in the key interactions and showed that this docking protocol was able to reproduce the correct pose (Fig 2).
Fig. 2.
Superimposition of the ADP-ribose after docking (in red, the top pose) on its structure in the co-crystal structure (in blue) at the active site of nsP3. The heavy-atom RMSD between them is about Å
Table 1.
Comparison of the hydrogen-bonding interactions inferred for the nsP3–ADP-ribose docked complex with previously published data. In [17], key residues were identified by performing an experimental study of the crystal structures of the complex nsP3–ADP-ribose (3GPO), while the residues reported in [16] were determined by realizing molecular dynamics simulations of ADP-ribose in nsP3, based on the above crystal structures
| From the present work | From [17] | From [16] | |
|---|---|---|---|
| HBs | 11 | 11 | 11 |
| Interacting residues | Ile11, Ala23, Asn24, Asp31, Val33, Leu108, Gly112, Val113, Tyr114, Tyr142, Arg144 | Asp10, Ile11, Asn24, Asp31, Thr111, Gly112, Val113, Tyr114, Tyr142, Arg144 | Asp10, Ile11, Asn24, Asp31, Val33, Ser110, Thr111 Gly112, Val113, Tyr114, Arg144 |
Identification of potential inhibitors using virtual screening
In an effort to identify potential inhibitors (hit compounds), 1541 compounds in NCI Diversity Set II were screened by docking them with the nsP3 of CHIKV. Virtual screening (VST) was carried out using three different setups. The first was a focused docking centered around the ADP-ribose binding site (pocket 1: VST1 and VST2). The second setup was a blind docking centered either at the middle of the ADP-ribose binding site (VST3) or the protein (VST4), with the box made large enough to cover the whole protein. The third setup was a focused docking centered on the binding sites predicted by MetaPocket (pocket 2: VST5; pocket 3: VST6). The top ten compounds for each VST based on their binding affinities are listed in Table S2 of the ESM.
In the focused dockings that targeted binding pocket 1 (the ADP-ribose binding site; VST1 and VST2), the top hits were NCI_25457 (−10.8 kcal mol−1) and NCI_345647_a (−10.9 kcal mol−1). Among the top ten hits, four are shared between VST1 and VST2, which differ in the size of the grid box used. Given our interest in finding more inhibitors as well as other potential binding sites in the structure of nsP3, we used blind docking to dock into the entire protein, with the grid box centered on the middle of either the ADP-ribose binding site (VST3) or the protein (VST4). For the blind docking (VST3 and VST4), six of the top ten hits are common to VST3 and VST4, and the binding affinities were reproduced to within 1.0 kcal mol−1. This indicates that the blind dockings are likely to have converged. The results show that most of the top ten hits fitted well in pocket 1, and this pocket can accommodate ligands of different sizes. However, ligands with bulky structures, such as NCI_293778, NCI_58052, and NCI_61610 (common to the top ten hits of both VST3 and VST4), protruded from the binding site. Furthermore, the other pockets surrounding pocket 1 may serve as alternative binding sites for potential inhibitors.
By comparing the results from the blind dockings with the pockets predicted for nsP3 using MetaPocket, pockets 2 and 3 were identified, and focused docking at pockets 2 (VST5) and 3 (VST6) was conducted. Screening at VST5 and VST6 produced hits that had already been identified in previous screens, along with some new hits (Table S2 of the ESM). Pockets 1 and 3 share a number of interacting residues, including Asn24, Asp31, Val33, Gly112, Val113, and Tyr114. In contrast, pocket 2 was located on the opposite side and behind pocket 1, and comprised the residues His1, Pro2, Ser3, Tyr4, Met132, Asp133, Ser134, Thr135, Asp136, Ala137, Asp138, Val139, Arg159, Thr160, Ile156, and Gln157. The locations of the pockets in nsP3 and the locations of the different ligands in the three pockets are illustrated in Figs. 3 and 4. Most of the compounds effectively occupy pockets 2 and 3 and participate in significant interactions. Ligands NCI_127133 (−8.3 kcal mol−1) and NCI_670283 (−10.6 kcal mol−1) bind in pockets 2 and 3 as well as pocket 1, but in different conformations. Interestingly, the ligand NCI_293778 would be able to bind into all three pockets, although it may be inferred that pocket 1 is more favorable for binding, with a binding affinity of −10.5 kcal mol−1 compared to −9.4 kcal mol−1 (pocket 3) and −8.3 kcal mol−1 (pocket 2).
Fig. 3a–b.
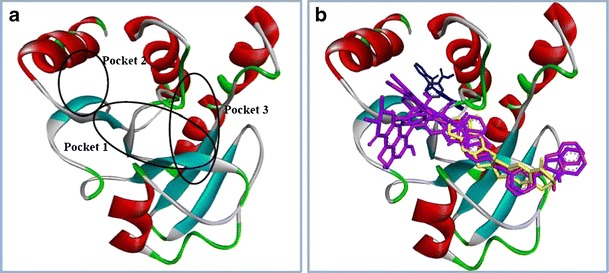
Representation of three binding pockets identified in nsP3. a The locations of three pockets in nsP3. b Illustration of the three pockets showing the locations of five top-hit compounds obtained from virtual screenings at these binding sites: NCI_25457, NCI_345647_a, NCI_61610 (in magenta) at pocket 1, NCI_127133 (in blue) at pocket 2, and NCI_670283 (in yellow) at pocket 3
Fig. 4a–c.
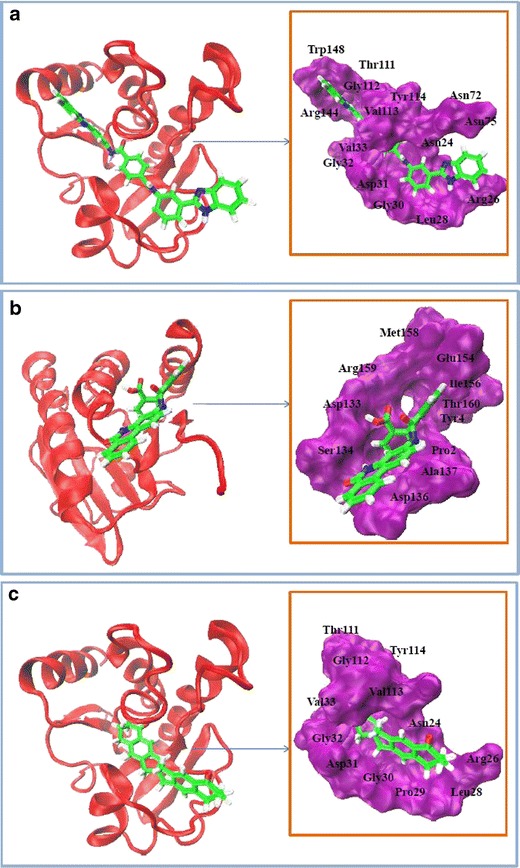
Illustrations of three binding pockets identified in nsP3. Each illustration contains two images: the first shows the protein nsP3 (in red) as a ribbon and the ligand (in green) in stick representation, and the second shows the surface of the pocket (in purple) surrounding the bound ligand (in green) in stick representation. a Pocket 1 with the complex nsP3–NCI_61610. b Pocket 2 with the complex nsP3–NCI_127133 (structure has been rotated 180°). c Pocket 3 with the complex nsP3–NCI_670283
For the virtual screening, it is worth mentioning that the changes in the size of the grid box and its location affected the search process in Vina. Increasing the dimensions of the box is likely to be better suited to larger molecules. For instance, in the blind docking (VST3 and VST4), NCI_61610 (−11.1 kcal mol−1) was identified as a top ten hit, but it is not one of the top ten hits for VST1 and VST2. The locations of the three binding sites along with the top five compounds obtained from VST are displayed in detail in Fig. 3 (for more information, see Fig. S1 of the ESM). It is also interesting that the majority of the hit compounds show tighter binding in pocket 1 than in pocket 2. Detailed analyses of the interactions between the ligands and the protein target were carried out by characterizing the hydrogen bonds and hydrophobic contacts [34]. The results show that hydrogen-bonding interactions play a more important role in the binding to pocket 1, whereas hydrophobic contacts are responsible for the interactions associated with the binding to pocket 2. In agreement with this, it was previously reported [16] that the residues Ser110, Thr111, Gly112, and Tyr114 define pocket 1 and are key residues in interactions with ligands. In addition, we found that these residues also define pocket 3.
Investigation into the stability of the protein and its complexes using molecular dynamics simulations
The top five hit compounds NCI_61610, NCI_25457, NCI_345647_a, NCI_670283, and NCI_127133 from each screening were subsequently subjected to molecular dynamics simulations (Table 2). MD simulations were undertaken to investigate the dynamic behavior of protein nsP3 and its complexes and to obtain the precise binding modes [35]. MD simulations were carried out with the NAMD package with the CHARMM force field for 20 ns following 3 ns of equilibrium simulations.
Table 2.
Chemical structures of the top five hit compounds and their properties
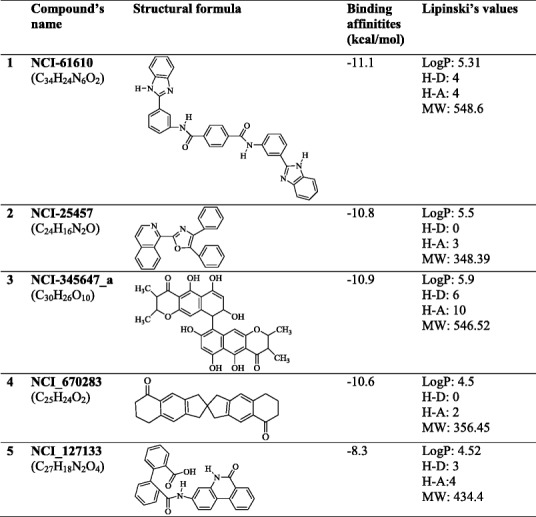
LogP an octanol-water partition coefficient; H-D hydrogen donor; H-A hydrogen acceptor; MW molecular weight
In order to assess the overall stability, RMSD values with respect to the starting structure were used as the major criterion. The backbone RMSD curves for nsP3 and its complexes with different ligands with respect to the starting structure were not significantly different from each other (Fig. 5). All systems were relatively stable throughout the simulations after reaching equilibrium within 3 ns. Hence, data taken from the trajectories in the last 20 ns will be used for further analysis.
Fig. 5.
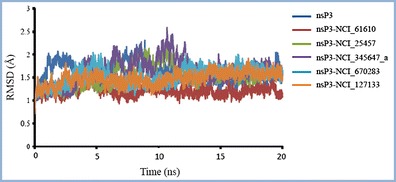
Backbone RMSD profiles for the apoprotein nsP3 and complexes of nsP3 with various ligands during MD simulations
The root mean square fluctuation (RMSF) of the Cα atom of each residue was calculated using the data from 20-ns trajectories of the protein nsP3 and its complexes (Fig. 6). The RMSF profiles of the apoprotein and the complexes were similar. However, subtle differences were observed for a few regions, including the loop at residues 31–34. The residues comprising the binding pockets were quite stable during the simulations, with fluctuations of <1.0 Å. It is worth noting that when the three ligands NCI_61610, NCI_25457, and NCI_345647_a were bound to the protein at pocket 1, the RMSFs for the binding loop region 31–34 were noticeably smaller than those seen for the apoprotein. The RMSF of this loop was not significantly perturbed when the ligands NCI_670283 and NCI_127133 were bound to pockets 3 and 2, respectively.
Fig. 6.
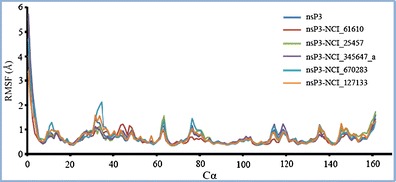
RMSF values of the Cα atoms of the apoprotein nsP3 and its different complexes, as calculated during MD simulations
Interactions between the ligands and the protein nsP3
A detailed analysis of the hydrogen-bonding (H-bond) interactions and hydrophobic contacts between the ligands and nsP3 was carried out. Simple geometric criteria were used to define a hydrogen bond: a distance between proton donor (D) and acceptor (A) atoms of <3.5 Å and an angle D–H…A of >120° [16]. If the H-bond occupation percentage was >50 %, they were considered to arise from the medium, whereas strong hydrogen bonds were considered to be present for H-bond occupations of greater than 75 % [16]. Hydrophobic contacts between the carbon atoms of nonpolar parts of protein residues were also monitored, with a cutoff distance of 4.0 Å assumed [36, 37].
Complementary to the results of the docking (where the protein was kept rigid), molecular dynamics simulations revealed that when the ligands bind to nsP3, the ligands or/and the residues in the binding pockets fluctuate, with the residues adapting their structures to better accommodate the ligands by forming H-bonds and/or hydrophobic contacts. This can be illustrated by superimposing the complex before the simulation (at 0 ns) on the complex after the simulation (at 23 ns); see Fig. 7. Not surprisingly, the figure shows that some residues, such as Val33, Val113, Tyr114, Arg144, and Trp148 (for ligands binding to pocket 1; see Fig. 7a, b, and c), display noticeable movement upon binding. For ligands NCI_127133 (pocket 2) and NCI_670283 (pocket 3), the results showed that the fluctuations of residues Ala1, Pro2, and Tyr4 (in pocket 2) and residues Ala22, Val33, Val113, and Tyr114 (pocket 3) were required for a correct fit into nsP3.
Fig. 7a–f.
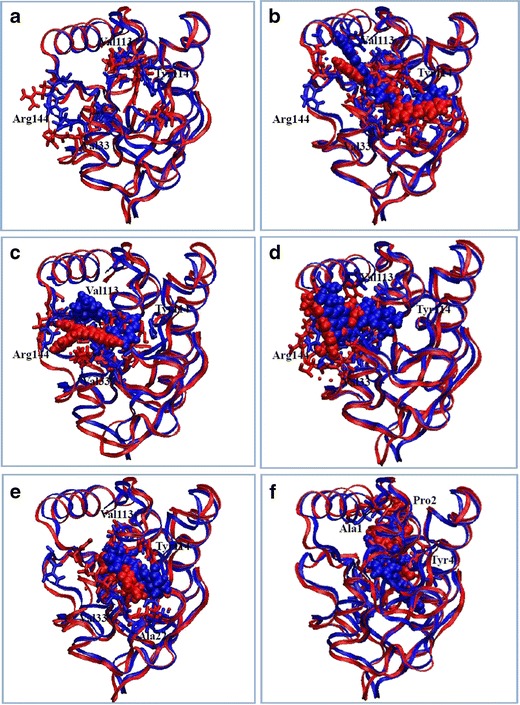
Superimposition of complex structures observed at 0 ns (in blue) and 23 ns (in red), with the structures represented as ribbons, the ligands as van der Waals surfaces, and the residues surrounding the ligands as sticks. a Apoprotein nsP3. b–f Complexes of nsP3 with the ligands NCI_61610 (b), NCI_25457 (c), NCI_345647_a (d), NCI_670283 (e), and NCI_127133 (f)
Key residues forming hydrogen bonds and hydrophobic contacts between nsP3 and ligands
Hydrogen-bonding analyses revealed that the binding of each investigated ligand to the protein is stabilized by a number of hydrogen bonds, except for ligand NCI_127133 (Table 3). We can infer that the binding site at pocket 1, where NCI_61610, NCI_25457, and NCI_345647_a bind, is the most favorable place for ligands to form hydrogen bonds (Fig. 8). There are three strong hydrogen bonds between ligand NCI_61610 and the residues Asn24 (98 %), Tyr114 (92 %), and Gly112 (88 %) in pocket 1, but only one strong and one moderately strong hydrogen bond between NCI_670283 and Thr111 (78 %) and Gly112 (66 %) in pocket 2 (Fig. 7). In contrast, pocket 3 is unlikely to be favored for binding, as no H-bonds with occupations of >10 % were identified.
Table 3.
Analysis of the hydrogen bonds that occurred during the trajectories sampled in the MD simulations
| Ligand | Number of H-bonds | H-bonds present | % Occupancy |
|---|---|---|---|
| NCI_61610 | 5 | Asn24 (HD22)–O1 | 98 |
| Tyr114 (HN)–O | 92 | ||
| Gly112 (HN)–O | 88 | ||
| Thr111 (OG1)–H1 | 13 | ||
| Cys34 (HG1)–O1 | 10 | ||
| NCI_25457 | 3 | Val113 (HN)–N | 29 |
| Val33 (HN)–N | 21 | ||
| Val33 (HN)–O | 20 | ||
| NCI_345647_a | 7 | Ile11 (HN)–O4 | 39 |
| Ile11 (HN)–O6 | 12 | ||
| Gly112 (HN)–O1 | 18 | ||
| Val33 (HN)–O | 17 | ||
| Gly32 (O)–O5 | 14 | ||
| Thr111 (HN)–O1 | 13 | ||
| Arg144 (HE)–O3 | 10 | ||
| NCI_670283 | 4 | Thr111 (HN)–O | 77 |
| Gly112 (HN)–O | 65 | ||
| Ser110 (HN)–O | 25 | ||
| Thr111 (HG1)–O | 10 | ||
| NCI_127133 | 0 | No H-bonds with occupancies of >10 % | |
Fig. 8a–b.
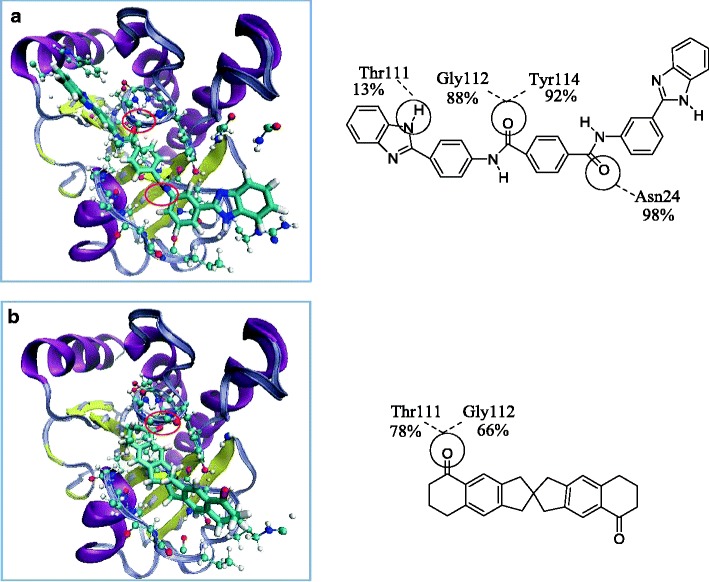
Hydrogen-bonding interactions between nsP3 and NCI_61610 and NCI_670283. a The complex of nsP3 with ligand NCI_61610 involves three strong H-bonds with residues Asn24, Tyr114, and Gly112. b The complex of nsP3 with ligand NCI_670283 involves one strong H-bond with Thr111 and one moderately strong H-bond with Gly112
Furthermore, the residues in the region 110–114 of the protein are key residues that are responsible for forming the hydrogen bonds between the protein and the ligands in binding pockets 1 and 3. Most of them serve as hydrogen-bond donors, except in the complex nsP3–NCI_61610, where residue Thr111 serves as an acceptor. This observation is in close agreement with the results of earlier simulations and experimental data [16]. In addition, it emerged that the structures of NCI_61610 and NCI_345647_a are more polar than the other ligands, so more hydrogen bonds were found in complexes involving them than in complexes involving the other ligands.
The nsP3 residues that were found to interact with the ligands through hydrophobic contacts are listed in Table 4. It is worth noting that four residues, namely Val33, Val113, Tyr114, and Trp148, play a crucial role in stabilizing the complexes by facilitating hydrophobic interactions between the ligands and nsP3 at binding pockets 1 and 3, while Ala1, Pro2, and Tyr4 are important in pocket 2. The solvent accessible surface areas (SASAs) were calculated to monitor the possible changes in the solvation environment upon ligand binding (Table 5). We expected to see that the SASA for hydrophobically interacting residues in the protein–ligand complex will decrease compared to those in the apoprotein. In pocket 1, when the ligands bind to the protein, it can be seen that the solvent surface of residue Tyr114 decreases from 70.2 Å2 in the apo nsP3 to 63.4 Å2 for the nsP3–NCI_61610 complex and 63.8 Å2 for the nsP3–NCI_345647_a complex. Also, the SASA of Val33 reduces from 68.8 Å2 in the apo state to 57.4 Å2 and 52.9 Å2 for the bound state in nsP3–NCI_25457 and NCI_345647_a, respectively. However, it is apparent that the changes in SASA for Val33, Val113, and Trp148 are not consistent for different ligands. This can be rationalized by noting that these residues are able to form not only hydrophobic contacts but also polar hydrogen-bonding interactions. Thus, the change in SASA is compromised by polar interactions, and both types of interaction modulate the SASA.
Table 4.
Analysis of hydrophobic contacts during the trajectories sampled in the MD simulations
| Ligand | Number of interacting residues | Interacting nonpolar residues |
|---|---|---|
| NCI_61610 | 8 | Ala22, Pro25, Leu28, Val33, Pro107, Val113, Tyr114, Trp148 |
| NCI_25457 | 6 | Ala22, Val33, Pro107, Val113, Tyr114, Trp148 |
| NCI_345647_a | 6 | Ile11, Val33, Ala36, Val113, Tyr114, Trp148 |
| NCI_670283 | 6 | Ala22, Leu28, Val33, Pro107, Val113, Tyr114 |
| NCI_127133 | 3 | Ala1, Pro2, Tyr4 |
Table 5.
SASA values obtained in the simulations of apo nsP3 and complexes of it with different ligands
| nsP3 | nsP3–NCI_61610 | nsP3–NCI_25457 | nsP3–NCI_345647_a | nsP3–NCI_670283 | nsP3–NCI_127133 | |
|---|---|---|---|---|---|---|
| Ala22 | 31.3 | 36.6 | 35.6 | 45.0 | ||
| Pro25 | 75.5 | 57.8 | ||||
| Leu28 | 102.6 | 108.2 | 100.8 | |||
| Val33 | 68.8 | 81.7 | 57.4 | 52.9 | 79.7 | |
| Pro107 | 25.9 | 20.4 | 25.2 | 24.1 | ||
| Val113 | 129.8 | 146.5 | 138.8 | 130.8 | 142.2 | |
| Tyr114 | 70.2 | 63.4 | 69.3 | 63.8 | 87.4 | |
| Trp148 | 84.0 | 88.4 | 90.2 | 95.5 | ||
| Ile11 | 12.6 | 25.8 | ||||
| Ala36 | 34.0 | 30.0 | ||||
| Ala1 | 74.5 | 81.2 | ||||
| Pro2 | 37.9 | 56.4 | ||||
| Tyr4 | 39.2 | 41.7 |
In order to probe the effects of the static protein structure used in the docking, multiple docking simulations and virtual screening were carried out based on the sampled conformations of nsP3–NCI_61610 at different simulation times (5, 10, 15, and 20 ns). These results are listed in Tables S3 and S4 of the ESM. The binding affinities observed in these docking runs (Table S3 of the ESM) were not significantly different from those obtained in previous dockings based on the X-ray structure. Additionally, the binding modes observed in the dockings based on the X-ray structure are similar to those obtained based on the MD sampled structures. Most of the top hits in the virtual screening were the same compounds as those identified in the previous screening, although there were some new hits too. This indicates that the docking results were similar for different static structures of the nsP3 protein.
Discussion and conclusions
So far, little information on the nsP3 macro domain and its inhibitors has been reported. The present study is of great importance in relation to identifying novel inhibitors of the chikungunya virus. It is also the first to combine molecular docking, virtual screening, and molecular dynamics simulations to identify potential inhibitors that target CHIKV by taking advantage of the recently determined high-resolution crystal structure of nsP3 in complex with ADP-ribose. Considering the presence of positive charge at the ADP-ribose-binding site, this binding pocket may have the tendency to accommodate negatively charged ligands, which may not have acceptable pharmacokinetic properties. Therefore, we decided to identify all possible binding pockets based on the available methods. We first re-docked the ADP-ribose and successfully reproduced the bound structure and the key residues for interactions in the ADP-ribose-binding pocket. It was evident that the binding affinity of ADP-ribose was less than −10 kcal mol−1, showing that this negatively charged ligand binds very tightly to nsP3. This study also demonstrates that the current docking protocol utilizing Autodock Vina is a robust approach for reproducing experimentally determined binding modes.
Subsequently, virtual screening with the NCI Diversity Set II was undertaken to identify potential novel inhibitors that target CHIKV. In addition to the well-characterized adenine binding pocket (pocket 1), two additional binding pockets (pockets 2 and 3) were identified through blind docking and using MetaPocket. Pocket 3 partially overlapped with pocket 1, while pocket 2 was located on the opposite side of the protein (Fig. 3). A comparison of the binding affinities at the three different binding pockets revealed that all of the hits for pocket 1 bind to the protein well, with binding affinities of less than −10 kcal mol−1. This suggests that pocket 1 is the most favorable for ligand binding. In contrast, pocket 2 may not be a good location for binding compared to pockets 1 and 3 because it is associated with higher binding affinities. It is also interesting to note that some ligands were repeated multiple times in the list of virtual screening results. These ligands can bind to different pockets in different conformations (e.g., the ligand NCI_127133 binds to pockets 1 and 2, ligand NCI_670283 binds to pockets 1 and 3, and ligand NCI_293778 binds to pockets 1, 2, and 3). These ligands may therefore have greater potential for use as inhibitors.
By analyzing the hydrogen bonding and hydrophobic interactions between the ligands and nsP3, key residues were identified. The region encompassing residues 110–114 was predicted to be the most important area for ligand interactions. This is consistent with the results of previous work by Rungrotmongkol et al. [16].
Furthermore, equilibrium molecular dynamics simulations were carried out to validate the molecular docking results. Analysis of the simulations confirmed that the docked ligand–nsP3 complexes were stable during the 20-ns simulation. However, subtle structural rearrangements in nsP3 were observed that led to better accommodation of the ligands. MD simulations also confirmed that binding pocket 1 is the favored pocket for ligand binding, considering the favorability of the interactions between the ligands and nsP3.
In summary, through a combination of molecular docking, virtual screening, and molecular dynamics simulations, we identified potential inhibitors of CHIKV based on the targeting of its nsP3 macro domain. In future work, the detailed binding modes of the identified inhibitors will be better characterized by performing more computationally extensive free-energy calculations, and their inhibitory effects will be verified using experimental studies. Additionally, the present work provides useful information for constructing pharmacophores in ligand-based drug design and for generating the chemical libraries utilized in the development of novel inhibitors targeting CHIKV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 1590 kb)
Acknowledgments
This project was funded by a University of Wollongong UOW-Vietnamese Government Scholarship (VIED-MOET). H.Y. is the recipient of an Australian Research Council Future Fellowship (project number FT110100034). This research was undertaken with the assistance of resources provided by the University of Wollongong High Performance Computing Cluster.
Footnotes
This paper belongs to a Topical Collection on the occasion of Prof. Tim Clark’s 65th birthday
Contributor Information
Haibo Yu, Email: hyu@uow.edu.au.
Paul A. Keller, Email: keller@uow.edu.au
References
- 1.Her Z, Kam Y-W, Lin RTP, Ng LFP. Chikungunya: a bending reality. Microbes Infect. 2009;11(14–15):1165–1176. doi: 10.1016/j.micinf.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Robinson MC. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955;49(1):28. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 3.Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88(Pt 9):2363–2377. doi: 10.1099/vir.0.82858-0. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8(7):491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler SA, Lu L, da Rosa AP, Xiao SY, Tesh RB. An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg. 2008;79(1):133–139. [PubMed] [Google Scholar]
- 6.Queyriaux B, Simon F, Grandadam M, Michel R, Tolou H, Boutin J-P. Clinical burden of chikungunya virus infection. Lancet Infect Dis. 2008;8(1):2–3. doi: 10.1016/S1473-3099(07)70294-3. [DOI] [PubMed] [Google Scholar]
- 7.Santhosh SR, Dash PK, Parida MM, Khan M, Tiwari M, Lakshmana Rao PV. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian Chikungunya virus isolates. Virus Res. 2008;135(1):36–41. doi: 10.1016/j.virusres.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Chandak NH, Kashyap RS, Kabra D, Karandikar P, Saha SS, Morey SH, Purohit HJ, Taori GM, Daginawala HF. Neurological complications of Chikungunya virus infection. Neurol India. 2009;57(2):177–180. doi: 10.4103/0028-3886.51289. [DOI] [PubMed] [Google Scholar]
- 9.Singh SK, Unni SK. Chikungunya virus: host pathogen interaction. Rev Med Virol. 2011;21(2):78–88. doi: 10.1002/rmv.681. [DOI] [PubMed] [Google Scholar]
- 10.Sourisseau M, Fsihi H, Frenkiel M-P, Blanchet F, Afonso PV, Ceccaldi P-E, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Schilte C, Saïb A, Rey FA, Arenzana-Seisdedos F, Desprès P, Michault A, Albert ML, Schwartz O, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost M-C. Characterization of reemerging chikungunya virus. Plos Pathog. 2007;3(6):e89–e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashad AA, Mahalingam S, Keller PA. Chikungunya virus: emerging targets and new opportunities for medicinal chemistry. J Med Chem. 2014;57:1147–1166. doi: 10.1021/jm400460d. [DOI] [PubMed] [Google Scholar]
- 12.Rashad AA, Keller PA. Structure based design towards the identification of novel binding sites and inhibitors for the chikungunya virus envelope proteins. J Mol Graph Model. 2013;44:241–252. doi: 10.1016/j.jmgm.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Voss J, Vaney MC, Duquerroy S, Vonrhein C, Ginard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468(7324):709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 14.Singh KD, Kirubakaran P, Nagarajan S, Sakkiah S, Muthusamy K, Velmurgan D, Jeyakanthan J. Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J Mol Model. 2012;18(1):39–51. doi: 10.1007/s00894-011-1018-3. [DOI] [PubMed] [Google Scholar]
- 15.Gould EA, Delogu I, Forrester N, Khasnatinov M, Gritsun T, de Lamballerie X, Canard B, Coutard B, Malet H, Morin B, Jamal S, Weaver S, Gorbalenya A, Moureau G, Baronti C. Understanding the alphaviruses: recent research on important emerging pathogens and progress towards their control. Antivir Res. 2010;87(2):111–124. doi: 10.1016/j.antiviral.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rungrotmongkol T, Nunthaboot N, Malaisree M, Kaiyawet N, Yotmanee P, Meeprasert A, Hannongbua S (2010) Molecular insight into the specific binding of ADP-ribose to the nsP3 macro domains of chikungunya and venezuelan equine encephalitis viruses: molecular dynamics simulations and free energy calculations. J Mol Graph Model 29(3):347–353. doi:10.1016/j.jmgm.2010.09.010 [DOI] [PubMed]
- 17.Malet H, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie X, Canard B, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol. 2009;83(13):6534. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaStarza MW, Lemm JA, Rice CM. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J Virol. 1994;68(9):5781–5791. doi: 10.1128/jvi.68.9.5781-5791.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dé I, Fata-Hartley C, Sawicki SG, Sawicki DL. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J Virol. 2003;77(24):13106–13116. doi: 10.1128/JVI.77.24.13106-13116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YF, Sawicki SG, Sawicki DL. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J Virol. 1994;68(10):6466–6475. doi: 10.1128/jvi.68.10.6466-6475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lastarza MW, Grakoui A, Rice CM. Deletion and duplication mutations in the C-terminal nonconserved region of sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology. 1994;202(1):224–232. doi: 10.1006/viro.1994.1338. [DOI] [PubMed] [Google Scholar]
- 22.Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385(1):212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Accelrys Software Inc. (2013) Discovery Studio modeling environment, 40th edn. Accelrys Software Inc., San Diego
- 25.Huang B. MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS. 2009;13(4):325–330. doi: 10.1089/omi.2009.0045. [DOI] [PubMed] [Google Scholar]
- 26.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 27.Phillips JC, Schulten K, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKerell AD, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Bashford D, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M, Bellott DRL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Woods RJ, Cheatham RTE, Darden T, Gohlke H, Luo R, Merz JKM, Onufriev A, Simmerling C, Wang B. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich E, Lalith P, Max LB, Tom D, Hsing L, Lee GP. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 31.Brooks BR, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Brooks RCL, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Mackerell JAD, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph Model. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Trott O, Olson AJ. Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badrinarayan P, Sastry GN. Virtual high throughput screening in new lead identification. Comb Chem High Throughput Screen. 2011;14(10):840–860. doi: 10.2174/138620711797537102. [DOI] [PubMed] [Google Scholar]
- 35.van Gunsteren WF, Hünenberger PH, Kastenholz MA, Oostenbrink C, Schenk M, Trzesniak D, van der Vegt NFA, Yu HB, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glättli A. Biomolecular modeling: goals, problems, perspectives. Angew Chem Int Ed Engl. 2006;45(25):4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 36.Bruno A, Guadix AE, Costantino G. Molecular dynamics simulation of the heterodimeric mGluR2/5HT(2A) complex. An atomistic resolution study of a potential new target in psychiatric conditions. J Chem Inf Model. 2009;49(6):1602–1616. doi: 10.1021/ci900067g. [DOI] [PubMed] [Google Scholar]
- 37.Rajgaria R, McAllister SR, Floudas CA. Towards accurate residue-residue hydrophobic contact prediction for alpha helical proteins via integer linear optimization. Proteins. 2009;74(4):929–947. doi: 10.1002/prot.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 1590 kb)


