Abstract
We previously reported that a high-mannose binding lectin KAA-2 from the red alga Kappaphycus alvarezii, which is an economically important species and widely cultivated as a source of carrageenans, had a potent anti-influenza virus activity. In this study, the full-length sequences of two KAA isoforms, KAA-1 and KAA-2, were elucidated by a combination of peptide mapping and cDNA cloning. They consisted of four internal tandem-repeated domains, which are conserved in high-mannose specific lectins from lower organisms, including a cyanobacterium Oscillatoria agardhii and a red alga Eucheuma serra. Using an Escherichia coli expression system, an active recombinant form of KAA-1 (His-tagged rKAA-1) was successfully generated in the yield of 115 mg per a litter of culture. In a detailed oligosaccharide binding analysis by a centrifugal ultrafiltration-HPLC method with 27 pyridylaminated oligosaccharides, His-tagged rKAA-1 and rKAA-1 specifically bound to high-mannose N-glycans with an exposed α1-3 mannose in the D2 arm as the native lectin did. Predicted from oligosaccharide-binding specificity, a surface plasmon resonance analysis revealed that the recombinants exhibit strong interaction with gp120, a heavily glycosylated envelope glycoprotein of HIV with high association constants (1.48–1.61 × 109 M−1). Native KAAs and the recombinants inhibited the HIV-1 entry at IC50s of low nanomolar levels (7.3–12.9 nM). Thus, the recombinant proteins would be useful as antiviral reagents targeting the viral surface glycoproteins with high-mannose N-glycans, and the cultivated alga K. alvarezii could also be a good source of not only carrageenans but also this functional lectin(s).
Keywords: Anti-viral lectin, HIV, Alga, Carageenophyta, Kappaphycus alvarezii
Introduction
Human immunodeficiency virus (HIV) infects CD4+ cells, where a viral envelope glycoprotein gp120 interacts with a receptor CD4 and a co-receptor CXCR4 or CCR5 of the host cells, and subsequent insertion of a viral envelope glycoprotein gp41 into the host cell membrane triggers the fusion followed by entry of the virus (Kilby and Eron 2003). gp120 is a highly glycosylated protein, as half of its total mass is accounted by about 24 N-linked glycans including complex and high-mannose types (Geyer et al. 1988). These densely attached N-glycans shield the viral surface proteins to escape from neutralizing antibody recognition (Wei et al. 2003; Bonomelli et al. 2011). On the other hand, carbohydrate-binding agents that target those glycans on the viral envelope exhibit potent anti-viral activities via interfering the interaction between gp120 and CD4 (Hansen et al. 1989; Balzarini et al. 1991; Charan et al. 2000; Dey et al. 2000; Swanson et al. 2010).
Several high-mannose N-glycan-binding lectins are newly found and marked as potential microbicides having anti-HIV activities, such as cyanobacterial lectins of cyanovirin-N (CV-N) from Nostoc ellipsosoprum (Boyd et al. 1997), scytovirin (SVN) from Scytonema varium (Bokesch et al. 2003) and MVL from Mycrosystis viridis (Bewley et al. 2004), a bacterial lectin of actinohivin (AH) from actinomycete Longispora albida (Chiba et al. 2001), and algal lectins of griffithsin (GRFT) from a red alga Griffithsia sp. (Mori et al. 2005) and BCA from a green alga Boodlea coacta (Sato et al. 2011b). These lectins show strong anti-HIV activities through their binding to mannoside structures on gp120. However, high-mannose recognition mode subtly differed among them, as well as the primary structures were clearly distinct (Sato et al. 2011b; Koharudin and Gronenborn 2014). High-mannose N-glycan structures are also present on the surfaces of the other enveloped viruses such as influenza virus, hepatitis C virus (HCV), human herpes virus (HHV), West Nile virus, severe acute respiratory syndrome-related coronavirus (SARS-CoV), and Ebola virus (Okuno et al. 1992; Vigerust and Shepherd 2007). This suggests that high-mannose-binding lectins could also inactivate the entries of those viruses into host cells (Barrientos et al. 2003; Helle et al. 2006; O’Keefe et al. 2003, 2010).
A lectin ESA-2 from a red alga Eucheuma serra (Kawakubo et al. 1997) was recently found to possess the unique binding specificity with the preferential affinity for high-mannose N-glycans bearing the nonreducing terminal α1-3 Man in D2 arm (Hori et al. 2007). Since then, several lectins with similar binding specificity were isolated from some lower organisms, including red algae (Kawakubo et al. 1999; Sato et al. 2011a; Hung et al. 2011, 2015), a cyanobacterium (Sato et al. 2000, 2007), and bacteria (Cumsky and Zusman 1979; Sato et al. 2012; Koharudin et al. 2012; Whitley et al. 2013). Interestingly, these lectins commonly had tandem-repeated structures of a similar domain composed of about 67 amino acids, and grouped into two types of four and two repeats (Hori et al. 2007; Sato et al. 2007; Sato and Hori 2009) as exemplified with ESA-2 (four repeats) and a lectin OAA (two repeats) from a freshwater cyanobacterium Oscillatoria agardhii. Thus, we demonstrated the occurrence of a new lectin family which is widely distributed in lower organisms, indicating that ESA-2 and OAA are orthologous to each other. Among them, OAA has been characterized in details. The structural analyses of OAA and its complexes with oligomannosides revealed that this lectin recognized a core unit, Manα1-6(Manα1-3)Manα1-6(Manα1-3)Manβ- of the branched mannoside in high-mannose N-glycan (Koharudin et al. 2011; Koharudin and Gronenborn 2011, 2014). This recognition mode of OAA was novel as the other high-mannose-binding lectins mostly recognized the nonreducing end mannoses. Potent anti-HIV activities of this lectin family, including ESA-2, OAA, Pseudomonas fluorescens agglutinin (PFA), Myxococcus xanthus agglutinin (myxobacterial hemagglutinin, MBHA) (Koharudin et al. 2012), and Burkholderia oklahomensis agglutinin (BOA) (Whitley et al. 2013) were represented as expected by their specific binding nature to high-mannose sugar moiety. Strong anti-influenza virus activities through binding to the envelope hemagglutinin protein were also shown in PFL (identical to PFA) (Sato et al. 2012) and KAA-2 isolated from a cultivated red alga Kappaphycus alvarezii (Sato et al. 2011a).
Algae belonging to genus Eucheuma and Kappaphycus are widely cultivated as food and carageenophyte. K. alvarezii cultivation was originally started in the Philippines in latter half of the 1960s using the local varieties selected from the wild (Doty 1973; Parker 1974; Bindu and Levine 2011). After four decades, the Kappaphycus became the most widely cultivated commercial eucheumoid (Bindu and Levine 2011) and has globally been introduced for cultivation and some experimental purposes in tropical and subtropical area, including Asia, Africa, the Pacific Island, and American countries (Ask et al. 2003; Bindu and Levine 2011). We recently reported that KAA-2 isolated from the cultivated sample of K. alvarezii preferentially recognized high-mannose-type oligosaccharides bearing an exposed α1-3 Man at the nonreducing end in D2 arm and inhibited infection of various influenza virus strains with EC50s of low nanomolar levels through direct binding to the viral envelope hemagglutinin protein, which has several high-mannose N-glycans (Sato et al. 2011a). The results also suggest that KAA-2 may belong to a new lectin family mentioned above. However, the primary structure of KAA is unknown yet as well as its anti-HIV activity. In this study, we elucidated the primary structure of KAA-2 using peptide mapping and complementary DNA (cDNA) cloning and prepared its active recombinants using an Escherichia coli expression system. The native and recombinant lectins were examined for sugar-binding specificity and anti-HIV activity, including binding nature to gp120.
Materials and Methods
Materials
K. alvarezii lectins KAA-1 and KAA-2 (previously declared as ECA-1 and ECA-2) and Eucheuma serra lectin ESA-2 were prepared as described previously (Kawakubo et al. 1997, 1999) and kept at −20 °C until use. The cultivated specimen of K. alvarezii was stored in RNAlater (Life Technologies, CA, USA) at −20 °C until RNA extraction. A vector pGEM-T Easy (Promega, WI, USA) and E. coli strain DH-5α were used for subcloning PCR-amplified products. An expression vector pET-28a(+) (Merck, Darmstadt, Germany) and an E. coli strain SHuffle T7 Express (New England Biolabs, MA, USA) were used for recombinant lectin preparation. Pyridylaminated (PA)-sugars were purchased from Takara Bio (Tokyo, Japan), except PA-derivatives of oligomannosides including mannotriose (Manα1-6(Manα1-3)Man-OH) and mannopentaose (Manα1-6(Manα1-3)Manα1-6(Manα1-3)Man-OH) which were previously prepared (Hori et al. 2007). A recombinant glycosylated HIV-1 IIIB gp120 (baculovirus) was purchased from ImmunoDiagnostics (MA, USA). Jurkat cells and T lymphocyte-derived cells were propagated in RPMI 1640 medium (Life Technologies) supplemented with 10 % fetal calf serum (Nichirei Biosciences, Tokyo, Japan).
Preparation of S-Pyridylethylated Lectins
KAA-2 and ESA-2 were subjected to S-pyridylethylation according to the method described previously (Hori et al. 2007). S-pyridylethylated (PE-) KAA-2 and PE-ESA-2 were purified by reverse-phase (RP-) HPLC on YMC-Pack PROTEIN-RP (6.0 × 250 mm, YMC, Kyoto, Japan). Briefly, after injection of sample to the column equilibrated with 10 % (v/v) acetonitrile in 0.05 % (v/v) trifluoroacetic acid (TFA), the column was washed with the same solvent for 10 min and then eluted with a linear gradient (10–70 %) of acetonitrile in 0.05 % TFA for 30 min at a flow rate of 1.0 ml/min. The eluate was monitored by absorbance at 280 nm (A280), and the active fractions were recovered. SDS-PAGE for purified proteins was performed using a 15 % (w/v) gel (Schägger and von Jagow 1987).
Trypsin Digestion and Separation of Peptides
One hundred micrograms each of PE-KAA-2 and PE-ESA-2 were digested with L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Roche Diagnostics, Basel, Switzerland) (enzyme/substrate = 1:50 (w/w)) in 1 % (w/v) ammonium hydrogen carbonate at 37 °C for 24 h. For the isolation of peptide fragments, each digest was separated by RP-HPLC on TSKgel ODS-120T column (4.6 × 250 mm, Tosoh, Tokyo, Japan). Briefly, the digest was applied to the column equilibrated with 5 % (v/v) acetonitrile in 0.1 % (v/v) TFA. The column was washed with the same solvent for 5 min and then eluted with a linear gradient (5–65 %) of acetonitrile in 0.1 % TFA for 60 min at a flow rate of 1.0 ml/min. The eluate was monitored by absorbance at 220 nm (A220), and the active fractions were recovered.
Molecular Weight Determination
The molecular masses of purified PE-KAA-2, PE-ESA-2, and their trypsin-digested peptides were determined by electron spray ionization (ESI)-mass spectrometry (MS) using Finnigan LCQ (Thermo Fisher Scientific, MA, USA). The masses of purified recombinants were determined by ESI-MS using LTQ Orbitrap XL (Thermo Fisher Scientific).
N-Terminal Amino Acid Sequencing
The N-terminal amino acid sequences of PE-KAA-2 and the peptide fragments produced by trypsin digestion of PE-KAA-2 were determined by a protein sequencer Procise 492 HT (Life Technologies).
cDNA Cloning of KAAs
Total RNA was extracted from 2.5 g of the RNAlater-treated alga of K. alvarezii using 25 ml of a Plant RNA Isolation Reagent (Life Technologies). Messenger RNA (mRNA) purification from the total RNA was performed using a NucleoTrap mRNA Purification Kit (Macherey-Nagel, Düren, Germany). Full-length cDNAs were synthesized from 50 ng of a mRNA using GeneRacer Kit (Life Technologies) according to the manufacturer’s instruction. The first PCR for rapid amplification of the cDNA 5′end (5′RACE) was performed with a 50 μl reaction mixture containing 5 μl of 10× Blend Taq buffer (Toyobo, Osaka, Japan), 10 nmol each of dNTP, 30 pmol of GeneRacer 5′ Primer (Life Technologies) (5′-CGACTGGAGCACGAGGACACTGA-3′), 250 pmol of a degenerated primer KAA_5′RACE_R1 (5′-ATIGGICCYTCICCYTTRTAYTGC-3′), which was designed from the partial amino acid sequence of KAA-2 predicted by peptide mapping, 1 μl of 10-fold diluted synthesized cDNA and 1.25 U of Blend Taq DNA polymerase (Toyobo). Thermal cycling conditions consisted of denaturation at 94 °C for 3 min, followed by 30 cycles consisting of denaturation at 94 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 1 min, and the final extension step at 72 °C for 5 min. The nested PCR was performed by the same method, except that 1 μl of a 100-fold diluted solution containing the first PCR product was used as a template, 10 pmol of GeneRacer 5′ Nested Primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and 250 pmol of a degenerated primer KAA_5′RACE_R2 (5′-AYTGRTTYTCIACRTTRTAIGTRTC-3′) as a primer pair, and that the annealing of the reaction was performed at 58 °C. Nested PCR products were subcloned into pGEM-T Easy vector (Promega). DNA sequencing was performed using BigDye Terminator Cycle Sequencing Kit Ver. 3.1 with ABI 3130xl DNA sequencer (Life Technologies).
3′RACE was performed by the same method of 5′RACE as described above, except that a primer pair of GeneRacer 3′ Primer (5′-GCTGTCAACGATACGCTACGTAACG-3′) and a degenerated primer KAA_3′RACE_F1 (5′-AYACITAYAAYGTIGARAAYCARTGGGG-3′) for the first PCR and GeneRacer 3′ Nested Primer (5′-CGCTACGTAACGGCATGACAGTG-3′) and a degenerated primer KAA_3′RACE_F2 (5′-ARTAYAARGGIGARGGICCIATHGG-3′) for the nested PCR were used. At last, the full-length cDNAs encoding two KAA isolectins were amplified using a high-fidelity DNA polymerase KOD FX Neo (Toyobo) with primer pairs of KAA1_5′End_F (5′-TATAGCTGAGTCAAGTTACACCAAC-3′) and KAA1_3′End_R (5′-AGAGGGTGATCACGTTTTTAC-3′) or KAA2_5′End_F (5′-ACCACCAGCACCGTGCTACTCGCT-3′) and KAA2_3′End_R (5′-GAATGTCCTTACGTGGCCTTTAC-3′), which were designed from the 5′ and 3′ terminal sequences of KAA cDNAs obtained by 5′ and 3′RACEs.
Sequence Data Processing
Homologous sequences were searched with the basic local alignment search tool (BLAST) program. Amino acid sequence comparison with homologous proteins from various organisms was performed using Clustal Omega (Sievers et al. 2011).
Construction of Expression Vector for Recombinant KAA-1
The recombinant KAA-1 gene was designed by back-translation from its amino acid sequence and synthesized by Integrated DNA Technologies (IA, USA). The KAA-1 coding region flanked by the restriction enzyme recognition sites was amplified by means of the PCR using a primer pair of rKAA1_F (5′-GATAGCTAGCGGCCGCTATACAGTTCAAAACC-3′) and rKAA1_R (5′-GATCCTCGAGTTAGCTCGCCACGCCTTTAAAG-3′), where underlined nucleotides represent the recognition sites for Nhe I and Xho I, respectively. The PCR using PrimeSTAR HS DNA Polymerase (Takara Bio) consisted of denaturation at 98 °C for 5 min, followed by 30 cycles consisting of denaturation at 98 °C for 10 sec, annealing at 60 °C for 5 s, and extension at 72 °C for 70 s, and the final extension step at 72 °C for 5 min. An amplified DNA fragment was subcloned into pET-28a(+) (Merck) to yield pET28a-rKAA1, which can express recombinant KAA-1 (rKAA-1) with the N-terminal hexa-His tag followed by the thrombin cleavage site (Fig. 1).
Fig. 1.
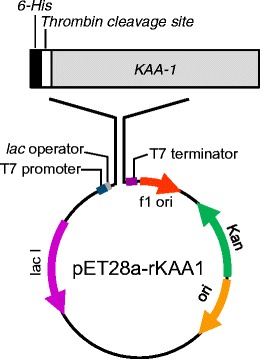
A diagram of recombinant KAA-1 (rKAA-1) expression vector pET28a-rKAA1. rKAA-1 was expressed as a 6-His- and thrombin cleavage site-fused protein
Expression of Recombinant KAA-1
E. coli SHuffle T7 Express strain (New England Biolabs), which is an enhanced BL21 derivative engineered to express a chromosomal copy of the disulfide bond isomerase DsbC (Chen et al. 1999) in the cytoplasm constitutively, was transformed with the rKAA-1 expression vector pET28a-rKAA1 by a conventional method. A transformant was cultured with 1 L of LB media at 37 °C until OD600 reached 0.5, and then, IPTG was added at a final concentration of 0.1 mM. After continuously cultured at 20 °C for 16 h, the bacteria cells were harvested by centrifugation at 10,000g for 20 min. After fracturing the cell by sonication, the His-tagged rKAA-1 (His-rKAA-1) expressed into soluble fraction was purified by His GraviTrap (GE Healthcare, Buckinghamshire, UK) according to the manufacturer’s instruction. After purification, His-rKAA-1 was dialyzed thoroughly against ultrapure water to remove imidazole which was contained in the elution buffer. Twenty-five milligrams of His-rKAA-1 was then applied to thrombin treatment using a Thrombin Cleavage Capture Kit (Merck) to disconnect the tag peptide from the recombinant according to the manufacturer’s instruction. After removal of thrombin by the above kit, the digested fraction was applied to His GraviTrap to exclude the cleaved His-tag peptide and undigested His-rKAA-1. Generated rKAA-1 was then dialyzed thoroughly against ultrapure water.
Measurement of Protein Concentrations
The A280 of solution was measured with NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Crude protein concentrations were represented as A280 at 1.0 mg/ml is 1.0. The purified protein concentrations were determined by calculating from A280 and their extinction coefficients which are computed from amino acid sequences (Gill and von Hippel 1989; Pace et al. 1995).
Hemagglutination Assay and Hemagglutination Inhibition Test
Hemagglutination assay was performed using a 2 % (v/v) suspension of trypsin-treated rabbit erythrocytes (TRBCs) as described previously (Hori et al. 1986). Hemagglutination activity was expressed as a titer, the reciprocal of the highest 2-fold dilution exhibiting positive hemagglutination. Hemagglutination-inhibition assay was also performed using a 2 % (v/v) suspension of TRBC and following sugars and glycoproteins as described in Hori et al. (1986); D-glucose, D-galactose, D-mannose, D-xylose, N-acetylglucosamine, N-acetylgalactosamine, N-acetylneuraminic acid, L-fucose, L-rhamnose, lactose, transferrin (human), asialo-transferrin, fetuin (type III, fetal calf serum), asialo-fetuin, yeast mannan, porcine thyroglobulin (PTG), asialo-PTG, bovine submaxillary mucin (BSM), and asialo-BSM. The inhibitory activity was expressed as the minimum concentration of sugar (mM) or glycoprotein (μg/ml) that completely inhibited the activity of a lectin solution of hemagglutination titer 8.
Centrifugal Ultrafiltration-HPLC Method for Determining Oligosaccharide Binding Specificity of rKAAs
The oligosaccharide binding activity of rKAAs was determined by a centrifugal ultrafiltration-HPLC method as described in Hori and Hirayama (2014). Briefly, 90 μl of 500 nM His-rKAA-1 or rKAA-1 in 50 mM Tris–HCl (pH7.0), and 10 μl of 300 nM PA-oligosaccharide were mixed and kept at room temperature for 60 min. Subsequently, unbound PA-oligosaccharides were collected by centrifugation (10,000g, 30 s) using Nonosep 10 K Device (Pall, NY, USA). An aliquot of the filtrate was applied to a TSKgel ODS 80TM column (4.6 × 150 mm, Tosoh), and unbound PA-oligosaccharide was quantified from the peak area. The amount of bound PA-oligosaccharide was obtained by subtracting the amount of unbound oligosaccharide from the amount of added oligosaccharide, which was determined from the filtrate of the reaction solution without a lectin. The binding activity (%) was calculated as the ratio of the amount of bound oligosaccharide to that of added oligosaccharide.
Surface Plasmon Resonance Analysis of Interaction of KAAs with HIV Envelope Protein gp120
Interaction of KAAs, including His-rKAA-1, rKAA-1, KAA-1, and KAA-2, with the HIV envelope gp120 was analyzed by surface plasmon resonance (SPR) analysis using a BIAcore 2000 system (GE Healthcare) as described previously (Sato et al. 2007, 2011b). The recombinant glycosylated HIV-1 IIIB gp120 (baculovirus) was immobilized to give 300 resonance units on a carboxymethylated dextran-coated sensor chip (CM5, GE Healthcare). Ninety microliters of each concentration of His-rKAA-1 and rKAA-1 solution (0.98, 3.9, 7.8, and 15.6 nM) was injected into the flow cells at 30 μl/min for 3 min to observe the association, and then, the same amount of the running buffer HBS EP (GE Healthcare) was injected into the flow cells at 30 μl/min for 3 min to observe the dessociation. The data of rKAAs were fit globally to a simple Langmuir 1:1 binding model with local Rmax (maximum response) using BIAevaluation 3.1 software. For KAA-1 and KAA-2, lectin solutions of 0.98 nM were applied to the system and the obtained sensorgrams were compared with those of the recombinants at same concentration to evaluate the functional similarities among the natives and recombinants.
Anti-HIV Activity
The HIV-1 virus stock was prepared from the Jurkat cells transfected with an HIV-1 genome-encoding plasmid, pNL4-3, originally constructed from the NY5 and RAV proviruses (Adachi et al. 1986). Infectivity of HIV-1 was measured with the median tissue culture infectious dose (TCID50) method using Jurkat cells. HIV-1 (100 TCID50) was incubated with 2-fold serially diluted lectins, in which their final concentrations were from 1 μM to 1 nM, at room temperature for 3 min in four or eight wells of a 96-well plate for each dilution. Jurkat cells (5 × 104 cells/well) were further added and incubated for several days until cytopathic effects developed. Fifty percent endpoint of lectin concentration to neutralize virus infectivity was determined according to the Behrens-Kaerber method, and 50 % inhibitory concentration (IC50) was determined. Cell viability in the presence of lectins was examined by using the Cell Proliferation Kit I (MTT assay; Roche Diagnostics, Tokyo, Japan).
Results
Peptide Mapping of KAA-2
Before peptide mapping, KAA-2 and ESA-2 were pyridylethylated and then purified as single peaks by RP-HPLC on YMC-Pack PROTEIN-RP column (Fig. 2a, b). The molecular masses of PE-KAA-2 and PE-ESA-2 were determined to be 28,124.0 and 28,052.0 by ESI-MS. These values were consistent with those estimated by SDS-PAGE (Fig. 2c). Peptide fragments generated by trypsin digestion of PE-KAA-2 and PE-ESA-2 were separated into a total of 26 peaks, including 18 common peaks between both, 5 specific peaks in PE-KAA-2, and 3 specific peaks in PE-ESA-2, by RP-HPLC on TSKgel ODS 120T (Fig. 3, Table 1). The molecular masses of the isolated peptide fragments were determined by ESI-MS, except for a peak 22 in PE-KAA-2 and peaks 10 and 21 in PE-ESA-2. The determined masses were compared to those of the peptide fragments that are theoretically generated by trypsin digestion of PE-ESA-2 (Table 1). All peptides whose masses were successfully determined were found in the PE-ESA-2 sequence, except for a peak 19, mass of which was not found in both sequences of PE-ESA-2 and trypsin used for digestion. The sequence of 167 amino acids (aa) of PE-KAA-2 was successfully predicted by comparing the sequences of 18 peptides with the same masses, which were commonly generated from both PE-KAA-2 and PE-ESA-2. Moreover, the N-terminal sequencing of the specific peptide peaks (11, 12, 15, 16, and 23) from PE-KAA-2 showed that there are at least 3 amino acid substitution for PE-ESA-2 (Fig. 4). The peptide sequence of a peak 10 (determined MW 2550.8) in PE-KAA-2 could be an incompletely digested product of 197GKLSGANNYSVENQWGGSSAPWNK220 (calculated MW 2551.7). Thus, a total of 230 aa in the whole sequence of PE-KAA-2 was successfully predicted by peptide mapping.
Fig. 2.
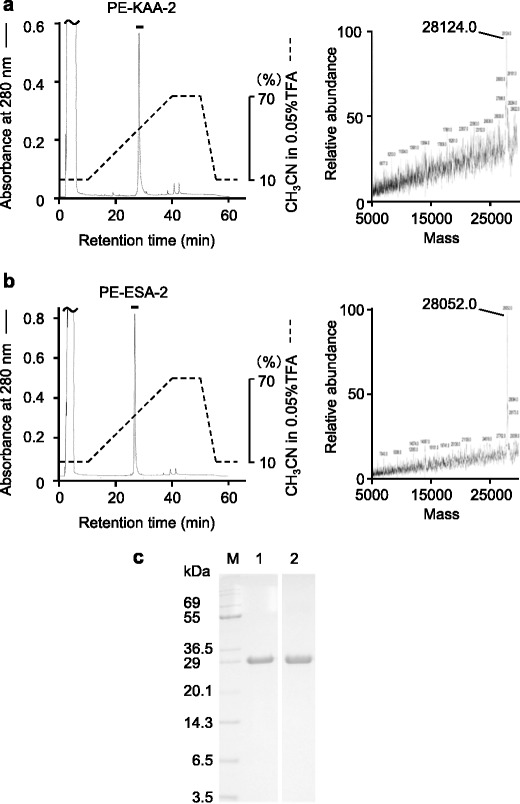
Isolation of PE-KAA-2 and PE-ESA-2 by reverse-phase HPLC. PE-KAA-2 (a) and PE-ESA-2 (b) were purified by RP-HPLC on YMC-Pack PROTEIN-RP column. The eluate was monitored by absorbance at 280 nm and the active fraction represented by a bar was recovered. Deconvoluted mass spectra of purified PE-KAA-2 (a) and PE-ESA-2 (b) using ESI-MS (LCQ) are also represented. c SDS-PAGE of PE-KAA-2 (lane 1) and PE-ESA-2 (lane 2). Five micrograms of protein was loaded in each lane. M molecular weight marker
Fig. 3.
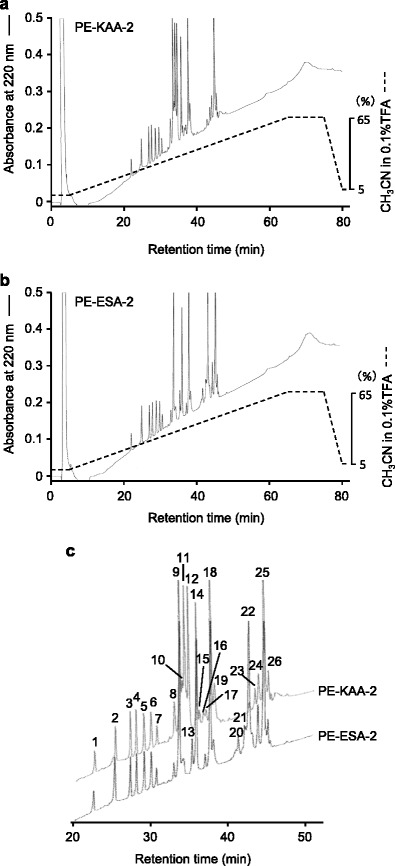
Peptide fragments generated by trypsin digestion of PE-KAA-2 and PE-ESA-2. Peptide fragments produced by trypsin digestion of PE-KAA-2 (a) and PE-ESA-2 (b) were purified by RP-HPLC on TSKgel ODS-120T column, and their enlarged images (c) are shown to compare the peak patterns. The peaks were numbered in serial order of retention time. Peaks 11, 12, 15, 16, and 23, and peaks 13, 20, and 21 were unique for PE-KAA-2 and PE-ESA-2, respectively
Table 1.
The molecular masses of peptide fragments produced by digesting PE-KAA-2 and PE-ESA-2 with trypsin and their corresponding sequence in ESA-2
| PE-KAA-2 | PE-ESA-2 | Sequence in ESA-2 | |||
|---|---|---|---|---|---|
| Peak no. | Determined molecular mass | Peak no. | Determined molecular mass | Calculated molecular mass | |
| 1 | 1023.6 | 1 | 1023.6 | 1024.1 | 230HNQNITAVK238 |
| 2 | 1275.4 | 2 | 1275.6 | 1276.3 | 247NLDGTCTYER256 |
| 3 | 1069.9 | 3 | 1069.5 | 1070.2 | 180TLEGTMQYK188 |
| 4 | 1171.4 | 4 | 1170.8 | 1172.3 | 113TLTGTMTYER122 |
| 5 | 746.2 | 5 | 746.3 | 746.9 | 123EGPIGFK129/257EGPIGFK263 |
| 6 | 831.3 | 6 | 831.3 | 831.9 | 189GEGPIGFR196 |
| 7 | 1588.7 | 7 | 1588.7 | 1589.7 | 96SGQGVLAVNITSSDGGK112 |
| 8 | 2539.5 | 8 | 2540.7 | 2540.6 | 130GTQSGGDTYNVENQWGGSSAPWNK153 |
| 9 | 2540.3 | 9 | 2539.5 | 2540.6 | 130GTQSGGDTYNVENQWGGSSAPWNK153 |
| 10 | 2550.8 | 10 | NDa | ||
| 11 | 2364.7 | –b | |||
| 12 | 1001.5 | – | |||
| – | 13 | 3403.5 | 3402.7 | 28GNQNVMAVDVNSSDGGANLNGTMTYSGEGPIGFK61 | |
| 14 | 957.4 | 14 | 957.4 | 958.1 | 154AGIWALGDR162 |
| 15 | 3490.5 | – | |||
| 16 | 3450.1 ± 3.96c | – | |||
| 17 | 3348.2 | 17 | 3348.5 | 3348.6 | 65RGESNVYDVENQWGGSSAPWHAGGQFVIGSR95 |
| 18 | 3347.3 | 18 | 3347.5 | 3348.6 | 65RGESNVYDVENQWGGSSAPWHAGGQFVIGSR95 |
| 19 | 3375.3 | 19 | 3375.9 | ||
| – | 20 | 3293.9 | 3293.5 | 199LSGANNYSVENQWGGSSAPWNAAGDWLIGDR229 | |
| – | 21 | ND | |||
| 22 | ND | 22 | 3293.0 | 3293.5 | 199LSGANNYSVENQWGGSSAPWNAAGDWLIGDR229 |
| 23 | 2759.8 ± 10.99c | – | |||
| 24 | 2763.3 | 24 | 2763.9 | 2764.0 | 3YTVQNQWGGSSAPWNDAGLWILGSR27 |
| 25 | 2763.2 | 25 | 2763.1 | 2764.0 | 3YTVQNQWGGSSAPWNDAGLWILGSR27 |
| 26 | 2800.2 | 26 | 2796.2 | 2764.0 | 3YTVQNQWGGSSAPWNDAGLWILGSR27 |
aNot determined
bNo consistent peak
cStandard error of >1.0 are represented
Fig. 4.
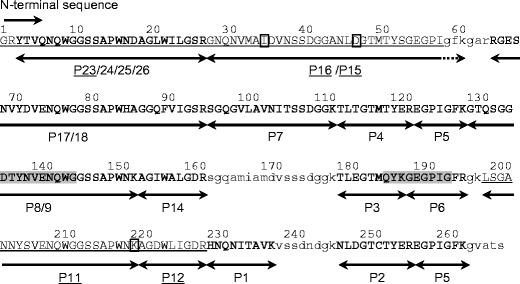
The predicted amino acid sequence of native KAA-2. The results of sequence prediction by peptide mapping in KAA-2 are shown using ESA-2 sequence as a template. The peptide fragments produced by trypsin digestion of PE-KAA-2 are represented with arrows. The peak numbers examined by Edman degradation and their determined sequences are underlined. The sequences whose peptide masses were consistent with those of ESA-2 are represented in bold. Unidentified amino acids in KAA-2 are represented as lowercase letter of corresponding sequence in ESA-2. Shaded regions in the sequence were used to design the degenerated primers for cDNA cloning
Nucleotide and Amino Acid Sequences of KAAs
cDNA cloning by RACE methods resulted in isolation of two full-length cDNAs encoding KAA isoforms (Fig. 5). Compared the deduced amino acid sequences of the two isoforms and the partial sequence of KAA-2 predicted by peptide mapping, it was found that there are substitutions of a few amino acids to one another, further indicating that KAA-2 isolated from the cultivated alga was not encoded by the two cloned cDNAs. This may be attributed to the difference of algal samples used for lectin isolation and cDNA cloning. Here, we defined the isolectin cDNA whose deduced amino acid sequence was closer to the predicted KAA-2 by peptide mapping as KAA-2 (DDBJ/EMBL/GenBank accession no. LC007081) and the other as KAA-1 (LC007080) (Fig. 5). KAA-1 cDNA was composed of 1027 bp containing 77 bp of 5′ untranslated region (UTR), 143 bp of 3′UTR, and 807 bp of open reading frame (ORF) which encoded 268 amino acids including an initiating methionine. KAA-2 cDNA was composed of 1161 bp containing 94 bp of 5′UTR, 257 bp of 3′UTR, and 810 bp of ORF encoding 269 amino acids including an initiating methionine (LC007081). The predicted N-terminal amino acid sequence of the mature KAAs was completely comparable in the sequence determined by Edman degradation for PE-KAA-2 (underlined sequence in Fig. 5a), indicating that KAA cDNAs do not encode signal peptide and other propeptide regions. These two KAAs showed 89.7 and 98.5 % identities to each other in nucleotide and protein levels, respectively. Amino acid sequence identities between ESA-2 and KAA-1 or KAA-2 are 98.1 and 98.9 %, respectively. KAAs also contain four tandem repeats of around 67 amino acids (Fig. 6) as well as ESA-2 (Hori et al. 2007). These repeated sequences are highly conserved between one another and among related proteins from other organisms including bacteria (Burkholderia oklahomensis agglutinin (BOA, 4 repeats), Myxococcus xanthus agglutinin (MBHA, 4 repeats), Pseudomonas fluorescens lectin (PFL, 2 repeats)) and cyanobacteria (O. agardhii (OAA, 2 repeats)) (Fig. 6). Similar sequence was not found in higher plants or animals by BLAST search. The sequence identities between KAA-1 and BOA, MBHA, OAA, or PFL were 58.1, 60.2, 60.6, and 62.9 %, respectively. The amino acids that interact with mannopentaose (Manα1-6(Manα1-3)Manα1-6(Manα1-3)Man), which is a branched oligomannosyl core structure of M8/9, in BOA (Koharudin et al. 2012) are absolutely conserved in all repeated domains of the lectins (indicated with asterisks in Fig. 6).
Fig. 5.
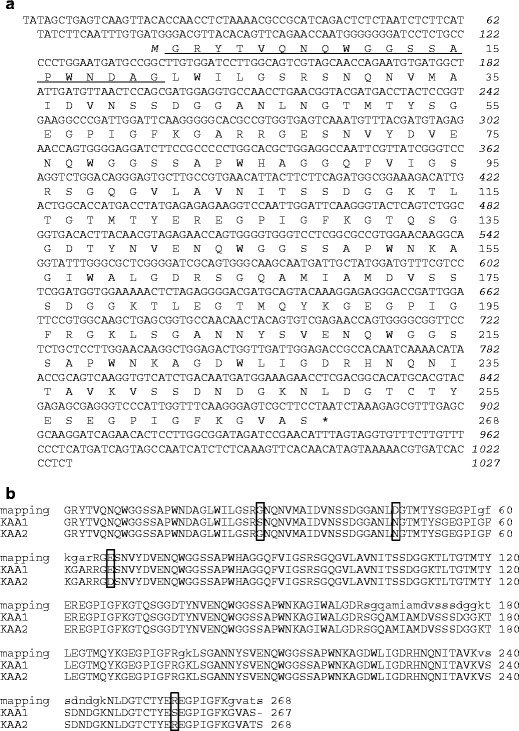
Nucleotide and deduced amino acid sequences of KAA-1. a Nucleotide and deduced amino acid sequences of the cDNA encoding KAA-1. The stop codon is shown as an asterisk. The italicized and nonitalicized numbers represent the positions of nucleotides and amino acids, respectively. Initiating methionine is indicated in italic. The 20 N-terminal amino acid sequence elucidated in Kawakubo et al. (1999) is underlined. b Comparison among deduced amino acid sequences of KAA-1 and KAA-2, and the sequence predicted by peptide mapping for native KAA-2 (see Fig. 2). Lowercase letters in native KAA-2 sequence represent the corresponding sequences in ESA-2. Amino acid substitutions among KAAs were boxed. The nucleotide sequence data of KAA-1 and KAA-2 appear in the DDBJ, EMBL, and GenBank databases under accession number LC007080 and LC007081, respectively
Fig. 6.
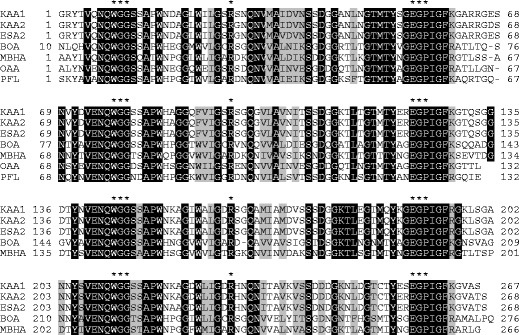
Alignment of amino acid sequences from KAAs and related proteins. Identical and similar residues among all proteins are indicated with black and gray backgrounds, respectively. Amino acids that interact with mannopentaose (Manα1-6(Manα1-3)Manα1-6(Manα1-3)Man) of M8/9 core unit in Burkholderia oklahomensis agglutinin (BOA) are indicated with asterisks. Data were cited from the following: Eucheuma serra agglutinin-2 (ESA-2, GenBank accession no. P84331), Burkholderia oklahomensis agglutinin (BOA, AIO69853), Myxococcus xanthus agglutinin (myxobacterial hemagglutinin (MBHA), M13831), Oscillatoria agardhii agglutinin (OAA, P84330), and Pseudomonas fluorescens lectin (PFL, ABA72252)
Preparation of Active Form of Recombinant KAA-1
His-rKAA-1 was successfully generated using pET system and a SHuffle T7 Express strain (Fig. 7). His-rKAA-1 was prepared in the yield of 115 mg per 1 l LB media, and rKAA-1 was generated in the yield of 15 mg from 25 mg of His-rKAA-1. The molecular masses of His-rKAA-1 and rKAA-1 were calculated as 30,202.7 and 28,451.9, respectively, based on their amino acid sequences. Although the recombinants ran at about 30–35 kDa in SDS-PAGE (Fig. 7a), the molecular masses determined by ESI-MS, 30,203.7 Da for His-rKAA-1 (Fig. 7b) and 28,452.1 Da for rKAA-1 (Fig. 7c), were in good agreement with the calculated ones. Hemagglutination actvities of His-rKAA-1 and rKAA-1 (each 10 μM) were 212 with TRBC as well as those of native KAA-1 and KAA-2 (data not shown).
Fig. 7.
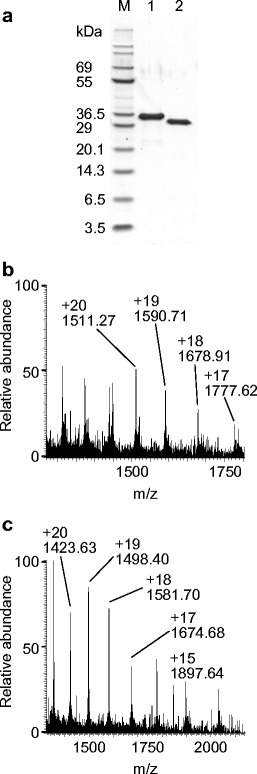
Preparation of rKAAs. a Nonreducing SDS-PAGE for purified His-rKAA-1 (lane 1) and rKAA-1 (lane 2). Five micrograms of each protein was loaded. M molecular weight marker. ESI-MS multiply charged spectra of purified His-rKAA-1 (b) and rKAA-1 (c) were shown
Carbohydrate and Oligosaccharide Binding Specificity of rKAAs
Hemagglutination inhibition test revealed that the recombinants (His-rKAA-1 and rKAA-1) and native KAA-1 and KAA-2 preferentially bound to glycoproteins containing high-mannose-type N-glycans such as yeast mannan and PTG, and weakly to the other glycoproteins without high-mannose N-glycan (Table 2). They had not affinity for any monosaccharide examined, including mannose. A relatively strong binding to BSM and asialo-BSM with O-glycans was also observed for the recombinants and native KAAs. Thus, the hemagglutination inhibition profiles of the recombinant KAAs were consistent with those of native KAAs, regardless of the presence or absence of the His-tag peptide in its N-terminus.
Table 2.
Hemagglutination-inhibition test of native and recombinant KAAs with sugar compounds
| Sugars and glycoproteins | Minimum inhibition concentration | |||
|---|---|---|---|---|
| KAA-1 | KAA-2 | His-rKAA-1 | rKAA-1 | |
| Monosaccharides and disaccharide (mM)a | >100 | >100 | >100 | >100 |
| Glycoproteins (mg/ml) | ||||
| N-glycan type | ||||
| High-mannose type | ||||
| Yeast mannan | 31.3 | 62.5 | 31.3 | 62.5 |
| High-mannose and complex types | ||||
| Porcine thyroglobulin (PTG) | 15.6 | <7.8 | 15.6 | 31.3 |
| Asialo-PTG | <7.8 | <7.8 | <7.8 | 15.6 |
| Complex type | ||||
| Transferin | >1000 | >1000 | 1000 | >1000 |
| Asialo-transferin | 500 | 250 | 250 | 500 |
| N/O-glycan type | ||||
| Fetuin | >1000 | >1000 | >1000 | >1000 |
| Asialo-fetuin | 250 | 250 | 250 | 500 |
| O-glycan | ||||
| Bovine submaxirally mucin (BSM) | 62.5 | 125 | 125 | 250 |
| Asialo-BSM | 31.3 | 31.3 | 62.5 | 125 |
aMonosaccharides (100 mM) (Glc, Man, Gal, GlcNAc, GalNAc, Rha, Fuc, Xyl, NeuAc) and a disaccharide (lactose) did not inhibit hemagglutination activities of four lectins
In oligosaccharide binding experiments by a centrifugal ultrafiltration-HPLC method with 27 PA-sugars (Figs. 8 and 9), His-rKAA-1 and rKAA-1 strongly bound to five oligosaccharides (oligosaccharides 7, 8, 10, 12 and 15) belonging to high-mannose-type N-glycans (oligosaccharides 7–17), but hardly to complex type N-glycans (oligosaccharides 1–6), oligomannoses (oligosaccharides 18–22), core structures (oligosaccharides 23 and 24), or oligosaccharides from glycolipids (oligosaccharides 25–27) (Fig. 9). Comparing the binding activities between oligosaccharides 7 (80.8 % in His-rKAA-1) and 16 (17.1 %), 8 (100 %) and 11 (19.3 %), 10 (100 %) and 13 (20.7 %), 12 (95.6 %) and 14 (36.7 %), or 15 (100 %) and 9 (12.7 %) revealed that rKAAs recognized the nonreducing terminal α1-3 linked Man of the D2 arm of branched oligomannoside, and the nonreducing terminal α1-2 linked Man in the D2 arm negatively affects for binding of rKAAs. The presence of His-tag hardly influenced to the oligosaccharide binding preferences of the recombinants although His-rKAA-1 and rKAA-1 were approximately 35 % apart in binding activities to oligosaccharides 7 and 17.
Fig. 8.
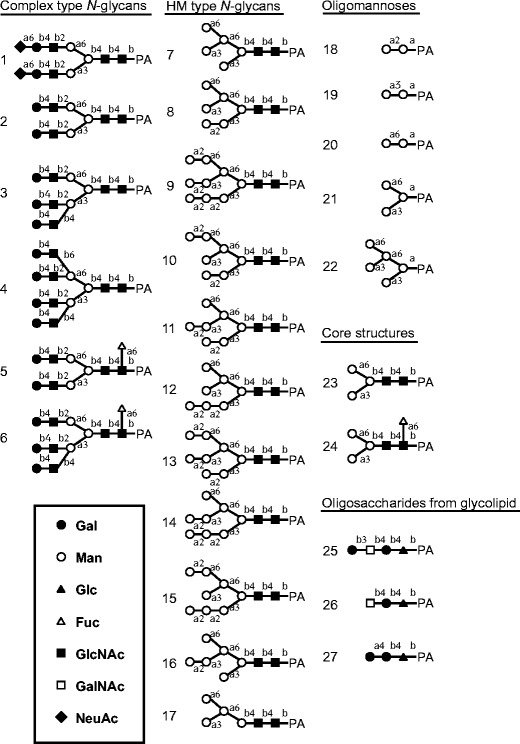
Structures of PA-oligosaccharides used in this study. Closed circle, galactose; open circle, mannose; closed triangle, glucose; open triangle, fucose; closed square, N-acetylglucosamine; open square, N-acetylgalactosamine; closed diamond, N-acetylneuraminic acid
Fig. 9.
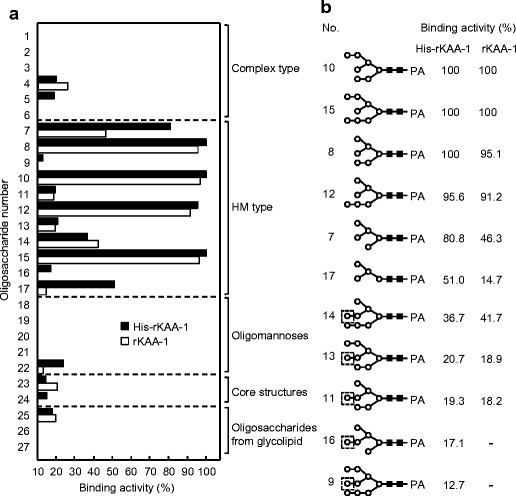
Interaction analysis between rKAAs and various oligosaccharides by a centrifugal ultrafiltration-HPLC method. a Binding activities of rKAAs to PA-oligosaccharides. Binding activity was expressed as a ratio (%) of the amount of a bound oligosaccharide to that of an added oligosaccharide. The assay was performed in duplicate for each PA-oligosaccharide, and the activity is expressed as the average value from duplicate assays. The activities less than 10 % were cut off for their insignificance. b The detailed binding activities of rKAAs with high-mannose-type N-glycans. Dashed boxes represent the nonreducing terminal α1-2 linked mannose residue in the D2 arm
Interaction of rKAAs with a Recombinant HIV Envelope Glycoprotein gp120
Interaction between rKAAs and HIV envelope glycoprotein gp120 was performed by SPR analysis using BIAcore2000 system. His-rKAA-1 and rKAA-1 bound to the gp120 immobilized on the chip dose-dependently (Fig. 10a, b) and binding kinetics of the interactions was well fitted to each sensorgram (overlaid red lines in Fig. 10a, b). rKAAs (Fig. 10a, b) showed high association constants (KA) (1.48–1.61 × 109 M−1) and low dessociation constants (KD) in the picomolar range (6.22–6.77 × 10−10 M) to HIV gp120, regardless of presence of His-tag (Table 3). The sensorgrams of KAAs including His-rKAA-1, rKAA-1, KAA-1, and KAA-2 at same concentration of 0.98 nM were very similar as shown in Fig. 10c. Although the resonance unit of the sensorgram in His-rKAA-1 appeared slightly higher than those of the other KAAs, it might be caused by the molecular weight difference, because the molecular weight of His-rKAA-1 having N-terminal His-tag peptide is approximately 2000 Da higher than those of the other KAAs.
Fig. 10.
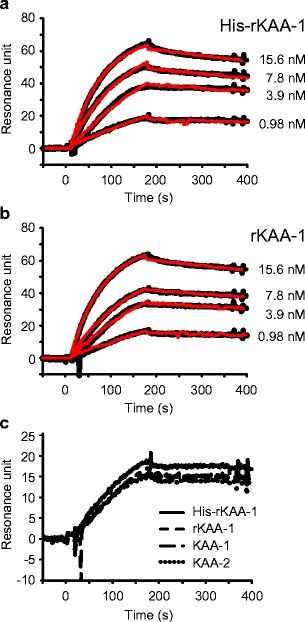
Interaction of KAAs with an HIV envelope glycoprotein gp120. SPR analyses for interaction between gp120 and His-rKAA-1 (a) or rKAA-1 (b) were analyzed with BIAcore2000. Each sensorgram represents the binding activity of rKAAs to gp120 on sensor chip. Ninety microliters of rKAAs solutions of each represented concentration were injected into the flow cells at 30 μl/min for 3 min. The binding response (black line) in resonance units is plotted against time (s). Binding kinetics of the interactions between rKAAs and gp120 (overlaid red line) were calculated by fitting the data to Langmuir model for 1:1 binding. c Comparison of binding activity to gp120 among KAAs including His-rKAA-1, rKAA-1, KAA-1, and KAA-2. SPR analysis was performed using 0.98 nM of each lectin solution by the same method as described above (color figure online)
Table 3.
Summary of binding kinetics of the interactions between rKAAs and gp120
| Lectin | ka (M−1s−1) | kd (s−1) | KA (M−1) | KD (M) |
|---|---|---|---|---|
| His-rKAA-1 | 8.71 × 105 | 5.90 × 10−4 | 1.48 × 109 | 6.77 × 10−10 |
| rKAA-1 | 8.50 × 105 | 5.29 × 10−4 | 1.61 × 109 | 6.22 × 10−10 |
k a association rate constant, k d dissociation rate constant, K A association constant, K D dissociation constant
Anti-HIV Activities of KAAs
In vitro anti-HIV activities of His-rKAA-1, KAA-1, and KAA-2 against HIV-1 strain (derived from pNL4-3) in Jurkat cells were determined by a neutralization assay. Serially diluted lectins and HIV-1 were reacted for 3 min at room temperature, and then, Jurkat cells were added and further incubated until cell fusion fully developed. Experiments were repeated at least three times for each lectin. Cell viability of Jurkat cells without HIV-1 was not affected by the same concentrations of lectins as judged by the MTT assay (data not shown). The results showed that KAAs prevented infection of HIV-1 at low nanomolar IC50 of 12.9, 9.2, and 7.3 nM for His-rKAA-1, KAA-1, and KAA-2, respectively (Table 4).
Table 4.
Neutralization of HIV-1 by native KAAs and a recombinant KAA-1
| Lectin | IC50 (nM) (± SD) |
|---|---|
| KAA-1 | 9.2 ± 2.2 |
| KAA-2 | 7.3 ± 1.9 |
| His-rKAA-1 | 12.9 ± 2.2 |
SD standard deviation
Discussion
Since Kawakubo et al. (1999) reported that the N-terminal amino acid sequences of KAAs were identical to those of ESAs, the whole primary structure of KAA-2 was considered to have high similarity with that of ESA-2. Peptide mapping of KAA-2 using the sequence of ESA-2 as a template has elucidated 85 % (230/268 aa) of the full-length sequence. A calculated molecular mass of the predicted sequence by peptide mapping, including the corresponding sequences of ESA-2, was 28,022, which gave close agreement with that of PE-KAA-2 (28,124 including an S-pyridylethyl group (105 Da) (Fig. 2a)). Although cDNA cloning by RACE method elucidated the full length cDNAs encoding KAAs, both of their deduced amino acid sequences were not identical to that of the native KAA-2 predicted by peptide mapping (Fig. 5b). The calculated molecular mass from their sequences were 27,881.24 and 28,007.41, respectively, and were also inconsistent with that of native KAA-2. We defined one of the two KAA cDNAs as KAA-2, whose amino acid sequence was more similar to native KAA-2 and ESA-2, and the other as KAA-1. The slight sequence differences between native KAA-2 and deduced KAA-2 might be individual variability in algal samples used. cDNA cloning has also elucidated that the transcripts of KAAs do not contain the region encoding a signal peptide sequence (Fig. 5), which target the proteins from the cytosol to the cytoplasmic membrane (prokaryotes) or to the endoplasmic reticulum membrane (eukaryotes) (Imai and Nakai 2010), as well as that of OAA from a cyanobacterium does not (Sato and Hori 2009). It has reported that cDNAs encoding a lectin from Euonymus europaeus (EEA) and EUL protein from Arabidopsis thaliana (ArathEULS3) do not encode a signal peptide and these translated product located in the nucleus and cytoplasm (Van Hove et al. 2011) as well as a GNA ortholog from rice that lacks the signal peptide and propeptide sequence (Fouquaert et al. 2007). In addition, there are not any known cellular localization signals in the primary structure of KAAs (data not shown). It suggests that KAAs are also synthesized on free ribosomes and located in nucleocytoplasm. There is only a report as to the cellular localization of KAA orthologs; Nelson et al. (1981) reported that MBHA, a KAA ortholog from a myxobacterium, localizes at the specific sites on the surface of developmental cells but not vegetated cells and suggested their function in cellular interactions during aggregation. Detailed cellular localization of KAA should be elucidated and will help to predict their functions in the algal bodies.
KAAs contain four tandem repeat domains in a molecule (Fig. 6). All the lectins of this family from algae are supposed to compose of four tandem repeat domains commonly, contrary to those from cyanobacteria and bacteria; in cyanobacteria, OAA composes of two repeats (Sato et al. 2007; Sato and Hori 2009) and a hypothetical protein from Lyngbya sp. (accession no. ZP_016222182) does four repeats (Sato and Hori 2009); in bacteria, PFL composes of two repeats (Sato et al. 2012) and BOA (Whitley et al. 2013) and MBHA (Koharudin et al. 2012) do four repeats (Fig. 6). Interestingly, several amino acid residues (23 aa per a single repeat domain) are absolutely conserved in all the domains among seven lectins shown in Fig. 6. The amino acid residues (7 aa per a single repeat domain) which interact with mannopentaose of a branched oligomannosyl core of M8/9 in BOA (Whitley et al. 2013) are also conserved (Fig. 6). This structural kinship suggested that lectins belonging to this lectin family might have similar carbohydrate-binding specificities.
A detailed oligosaccharide binding analysis by the centrifugal ultrafiltation-HPLC method using PA-sugars revealed that the recombinants, His-rKAA-1 and rKAA-1, preferentially recognized exposed α1-3Man in the D2 arm of high-mannose N-glycans (Figs. 8 and 9) as previously reported for native KAA-2 (Sato et al. 2011a). The nonreducing terminal α1-2 linked Man in the D2 arm tended to more negatively affect for binding to rKAAs than native KAA-2. The recognition mode of KAAs is absolutely different from the other high-mannose specific antiviral lectins including CV-N, GRFT, SVN, MVL, and AH (Koharudin and Gronenborn 2014). We also reported the same oligosaccharide binding properties in ESA-2 (Hori et al. 2007) and OAA (Sato et al. 2007), and recently in KSA from K. striatum (Hung et al. 2011) and EDA from E. denticulatum (Hung et al. 2015). A lectin PFL from Pseudomonas fluorescens Pf0-1 belonging to this lectin family also tends to preferentially recognize high-mannose glycans with α1-3 Man exposed in the D2 arm but not to other sugar types including complex N-glycans and O-glycans, as revealed by screening with a glycan array having 611 glycan targets (Sato et al. 2012). Three-dimentional structure analyses for glycan-bound OAA (Koharudin et al. 2011; Koharudin and Gronenborn 2011), PFA (identical to PFL), MBHA (Koharudin et al. 2012) and BOA (Whitley et al. 2013) supported their strict binding specificities; the amino acid residues which were revealed to interact with branched mannoside in the high-mannose glycans by tertiary-structural analyses were absolutely conserved in all the lectins of this family as mentioned above (Fig. 6). In previous (Sato et al. 2011a) and present studies, native KAA-2 and rKAAs bound to high-mannose-type oligosaccharides but not or little to oligomannoses including PA-mannopentaose (Manα1-6(Manα1-3)Manα1-6(Manα1-3)Man-PA) (oligosaccharide 22 in Fig. 8), suggesting a possibility that the reducing terminal disaccharide GlcNAc-GlcNAc is necessary for KAAs to bind to high-mannose N-glycans. On the other hand, Koharudin and Gronenborn (2011) showed that OAA binds to this type of mannopentaose (not pyridylaminated), by NMR binding studies, and suggested that the reducing terminal disaccharide was not essential for OAA to bind to those glycans. Exactly, PA-oligosaccharides used in this study possessed the reducing terminal residues with ring-opening structures by pyridylamination, including free oligomannoses (PA-oligosaccharides 18–22 in Fig. 8). Thus, it might be attributable to such ring-opening of the reducing terminal residues that KAAs did not bind to the PA-oligomannoses, including mannopentaose (Manα1-6(Manα1-3)Manα1-6(Manα1-3)Man-PA) (oligosaccharide 22 in Figs. 8 and 9). Although we have not elucidated the structure of a KAA-mannopentaose complex, it is strongly predicted that KAA, as well as OAA (Koharudin et al. 2011), requires the reducing terminal mannose having the ring structure for binding to mannopentaose, unlike that CV-N and GRFT do not directly interact with the reducing terminal mannose (Ziolkowska et al. 2006; Ziolkowska and Wlodawer 2006).
SPR analyses revealed that rKAAs bound to HIV envelope glycoprotein gp120 with high association constants (1.48–1.61 × 109 M−1) as well as native KAAs did (Fig. 10). gp120 is extensively glycosylated with N-linked complex and high-mannose carbohydrates accounting for about half of its molecular weight (Geyer et al. 1988), suggesting that KAAs can bind gp120 with a high affinity for the high-mannose sugars. Supposed from their strong binding to the gp120, KAAs represented potent anti-HIV activities at IC50 of nanomolar levels (7.3–12.9 nM, Table 4) in virus neutralization assay with Jurkat cells. The activities were comparable to those of OAA (12 nM of IC50 against HIV-1 (R9) in TZM-b1 cells (Koharudin et al. 2012), 45 nM of EC50 against HIV-1 X4 in MT-4 cells (Sato et al. 2007)), MBHA (15 nM of IC50 against HIV-1 (R9) in TZM-b1 cells (Koharudin et al. 2012)), BOA (12.2 nM of IC50 against HIV-1 (R9) in TZM-b1 cells (Whitley et al. 2013)) and ESA-2 (165 nM of EC50 against HIV-1 X4 in MT-4 cells (Sato et al. 2007)). There are a lot of HIV-1 strains, being suggested that there may be distinct sugar structures on their envelope gp120 (Zhang et al. 2004). The difference of anti-HIV activities, such as the activities of OAA against HIV-1 (R9) in TZM-b1 and HIV-1 X4 in MT-4 cells, may be caused by the difference of virus strains and cells (Huskens and Schols 2012) used in the experiments. The anti-HIV activities of CV-N and GRFT were reported as 0.4 nM (against HIV-1 (R9) in TZM-b1 cells) and 0.04 nM (against HIV-1 X4 in MT-4 cells) of IC50 (or EC50), respectively (Koharudin et al. 2012; Mori et al. 2005). The interaction between GRFT and HIV-1 gp120 IIIB (X4) was also elucidated by Xue et al. (2013) via a SPR analysis using a BIAcore T200; the dissociation constant of GRFT with the gp120 is 6.93 × 10−11 M, which is 10-fold lower than those of KAAs (6.22–6.77 × 10−10 M) (Fig. 10). These relatively strong anti-HIV-1 activities of CV-N and GRFT and the robust interaction of GRFT with gp120 come from the multivalency of binding sites on the lectins and that of epitopes on the high-mannose oligosaccharides (Koharudin and Gronenborn 2014). Although the anti-viral activities of KAAs are lower than those of CV-N and GRFT, the oligomannose epitopes recognized by KAAs are different from those recognized by CV-N and GRFT. Since high mutation rate in HIV-1 can rapidly produce the resistant viruses against these anti-viral lectins (Witvrouw et al. 2005; Huang et al. 2011), multilateral and synergistic use of the anti-viral lectins whose recognition epitopes in oligosaccharides are different from one another can be effective as Férir et al. (2014) represented.
We searched higher plant lectins similar to KAAs by in silico screening for genome sequences of soybean Glycine max, rice Oryza sativa, and Arabidopsis; however, higher plant genomes encode no lectins similar to this lectin family (data not shown) as Jiang et al. (2010) reported a genome wide screening of higher plant lectins. This lectin family is found to date only in lower organisms including eukaryotes of red algae and prokaryotes of cyanobacteria and bacteria. The biofunctional mechanism(s) where this lectin family preferentially binds to high-mannose carbohydrates having an exposed α1-3 Man in the D2 arm of branched oligomannoside is still uncovered. As Yoshiie et al. (2012) reported that the N-glycans of glycoproteins in red algae are predominantly high-mannose types, the internal ligands of KAAs could exist in the cells, implied from the predicted nucleocytoplasmic localization of the lectins as described before. However, the lectins peculiar to these lower organisms may have common function(s), such as host defense against infections of invasive viruses, bacteria, or fungi to these organisms as well as several cytoplasmic/nuclear plant lectins, including amaranthins, Euonymus lectin-like (EUL)-related lectins, jacalin-related lectins, and nictaba-related lectins, have an important role in plant defense against pathogens (Lannoo and Van Damme 2010, 2014; Dias Rde et al. 2015; De Schutter and Van Damme 2015).
This study showed that the lectins from the cultivated alga K. alvarezii inhibit the HIV virus entry to the host cells through binding to the virus envelope glycoprotein gp120 and confirmed their activities using its recombinants. These results suggested that KAAs have potentiality to represent broad anti-viral activities against viruses possessing high-mannose glycans on their envelope such as HCV, HHV, SARS-CoV, and Ebola virus, as well as native KAA-2 did against influenza viruses (Sato et al. 2011a). It is important that anti-viral lectins are isolated from a globally cultivated edible algal species in contrast that any other homologous anti-HIV lectins are derived from bacteria (BOA, PFL, MBHA), a cyanobacterium (OAA), and an uncultivated alga (ESA-2); KAAs can be purified in a relatively high yield of 270 mg per 100 g of freeze-dried alga (Kawakubo et al. 1999). These functional lectins will become a strong tool by being supplied in bulk as both native and recombinant forms, which are almost identical in their carbohydrate specificities and anti-viral activities.
Acknowledgments
Jurkat cells and pNL4-3 were kindly provided by Dr. Tsutomu Murakami in AIDS Research Center, National Institute of Infectious Diseases, Japan. We thank Dr. Tomoko Amimoto in the Natural Science Center for Basic Research and Development (N-BARD) at Hiroshima University for measuring the ESI-MS data. This work was partially supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry of Japan.
Compliance with Ethical Standards
Conflict of Interest
No conflicts of interest are declared.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ask EI, Batibasaga A, Zertuche-Gonzalez JA, de San M (2003) Three decades of Kappaphycus alvarezii (Rhodophyta) introduction to non-endemic locations. In: Chapman ARO, Anderson RJ, Vreeland VJ, Davison IR (eds) Proceedings of the 17th international seaweed symposium, Cape Town, South Africa, pp 49–57
- Balzarini J, Schols D, Neyts J, Van Damme E, Peumans W, De Clercq E. Alpha-(1–3)- and alpha-(1–6)-D-mannose-specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob Agents Chemother. 1991;35:410–416. doi: 10.1128/AAC.35.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos LG, O’Keefe BR, Bray M, Sanchez A, Gronenborn AM, Boyd MR. Cyanovirin-N binds to the viral surface glycoprotein, GP1,2 and inhibits infectivity of Ebola virus. Antiviral Res. 2003;58:47–56. doi: 10.1016/S0166-3542(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Cai M, Ray S, Ghirlando R, Yamaguchi M, Muramoto K. New carbohydrate specificity and HIV-1 fusion blocking activity of the cyanobacterial protein MVL: NMR, ITC and sedimentation equilibrium studies. J Mol Biol. 2004;339:901–914. doi: 10.1016/j.jmb.2004.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindu MS, Levine IA. The commercial red seaweed Kappaphycus alvarezii-an overview on farming and environment. J Appl Phycol. 2011;23:789–796. doi: 10.1007/s10811-010-9570-2. [DOI] [Google Scholar]
- Bokesch HR, O’Keefe BR, McKee TC, Pannell LK, Patterson GM, Gardella RS, Sowder RC, 2nd, Turpin J, Watson K, Buckheit RW, Jr, Boyd MR. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry. 2003;42:2578–2584. doi: 10.1021/bi0205698. [DOI] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O’Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH, 2nd, Buckheit RW, Jr, Nara PL, Pannell LK, Sowder RC, 2nd, Henderson LE. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charan RD, Munro MH, O’Keefe BR, Sowder RCII, McKee TC, Currens MJ, Pannell LK, Boyd MR. Isolation and characterization of Myrianthus holstii lectin, a potent HIV-1 inhibitory protein from the plant Myrianthus holstii. J Nat Prod. 2000;63:1170–1174. doi: 10.1021/np000039h. [DOI] [PubMed] [Google Scholar]
- Chen J, Song JI, Zhang S, Wang Y. Chaperon activity of DsbC. J Biol Chem. 1999;274:19601–19605. doi: 10.1074/jbc.274.28.19601. [DOI] [PubMed] [Google Scholar]
- Chiba H, Inokoshi J, Okamoto M, Asanuma S, Matsuzaki K, Iwama M, Mizumoto K, Tanaka H, Oheda M, Fujita K, Nakashima H, Shinose M, Takahashi Y, Omura S. Actinohivin, a novel anti-HIV protein from an actinomycete that inhibits syncytium formation: isolation, characterization, and biological activities. Biochem Biophys Res Commun. 2001;282:595–601. doi: 10.1006/bbrc.2001.4495. [DOI] [PubMed] [Google Scholar]
- Cumsky M, Zusman DR. Myxobacterial hemagglutinin: a development-specific lectin of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979;76:5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Van Damme EJM. Protein-carbohydrate interactions as part of plant defense and animal immunity. Molecules. 2015;20:9029–9053. doi: 10.3390/molecules20059029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Lerner DL, Lusso P, Boyd MR, Elder JH, Berger EA. Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses. J Virol. 2000;74:4562–4569. doi: 10.1128/JVI.74.10.4562-4569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Rde O, Machado Ldos S, Migliolo L, Franco OL. Insights into animal and plant lectins with antimicrobial activities. Molecules. 2015;20:519–541. doi: 10.3390/molecules20010519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty MS. Farming the red seaweed, Eucheuma, for carrageenans. Micronesia. 1973;9:59–73. [Google Scholar]
- Férir G, Huskens D, Noppen S, Koharudin LM, Gronenborn AM, Schols D. Broad anti-HIV activity of the Oscillatoria agardhii agglutinin homologue lectin family. J Antimicrob Chemother. 2014;69:2746–2758. doi: 10.1093/jac/dku220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquaert E, Hanton SL, Brandizzi F, Peumans WJ, Van Damme EJ. Localization and topogenesis studies of cytoplasmic and vacuolar homologs of the Galanthus nivalis agglutinin. Plant Cell Physiol. 2007;48:1010–1021. doi: 10.1093/pcp/pcm071. [DOI] [PubMed] [Google Scholar]
- Geyer H, Holschbach C, Hunsmann G, Schneider J. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem. 1988;263:11760–11767. [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hansen JE, Nielsen CM, Nielsen C, Heegaard P, Mathiesen LR, Nielsen JO. Correlation between carbohydrate structures on the envelope glycoprotein gp120 of HIV-1 and HIV-2 and syncytium inhibition with lectins. AIDS. 1989;3:635–641. doi: 10.1097/00002030-198910000-00003. [DOI] [PubMed] [Google Scholar]
- Helle F, Wychowski C, Vu-Dac N, Gustafson KR, Voisset C, Dubuisson J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J Biol Chem. 2006;281:25177–25183. doi: 10.1074/jbc.M602431200. [DOI] [PubMed] [Google Scholar]
- Hori K, Hirayama M. Centrifugal ultrafiltration-HPLC method for interaction analysis between lectins and sugars. In: Hirabayashi J, editor. Lectins. NY: Springer New York; 2014. pp. 173–183. [DOI] [PubMed] [Google Scholar]
- Hori K, Miyazawa K, Ito K. Preliminary characterization of agglutinins from seven marine algal species. Bull Jpn Soc Sci Fish. 1986;52:323–331. doi: 10.2331/suisan.52.323. [DOI] [Google Scholar]
- Hori K, Sato Y, Ito K, Fujiwara Y, Iwamoto Y, Makino H, Kawakubo A. Strict specificity for high-mannose type N-glycans and primary structure of a red alga Eucheuma serra lectin. Glycobiology. 2007;17:479–491. doi: 10.1093/glycob/cwm007. [DOI] [PubMed] [Google Scholar]
- Huang X, Jin W, Griffin GE, Shattock RJ, Hu Q. Removal of two high-mannose N-linked glycans on gp120 renders human immunodeficiency virus 1 largely resistant to the carbohydrate-binding agent griffithsin. J Gen Virol. 2011;92:2367–2373. doi: 10.1099/vir.0.033092-0. [DOI] [PubMed] [Google Scholar]
- Hung LD, Sato Y, Hori K. High-mannose N-glycan-specific lectin from the red alga Kappaphycus striatum (carrageenophyte) Phytochemistry. 2011;72:855–861. doi: 10.1016/j.phytochem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Hung LD, Hirayama M, Ly BM, Hori K. Purification, primary structure, and biological activity of the high-mannose N-glycan-specific lectin from cultivated Eucheuma denticulatum. J Appl Phycol. 2015;27:1657–1669. doi: 10.1007/s10811-014-0441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huskens D, Schols D. Algal lectins as potential HIV microbicide candidates. Mar Drugs. 2012;10:1476–1497. doi: 10.3390/md10071476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Nakai K. Prediction of subcellular locations of proteins: where to proceed? Proteomics. 2010;10:3970–3983. doi: 10.1002/pmic.201000274. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Ma Z, Ramachandran S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol Biol. 2010;10:79. doi: 10.1186/1471-2148-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakubo A, Makino H, Ohnishi J, Hirohara H, Hori K. The marine red alga Eucheuma serra J. Agardh, a high yielding source of two isolectins. J Appl Phycol. 1997;9:331–338. doi: 10.1023/A:1007915006334. [DOI] [Google Scholar]
- Kawakubo A, Makino H, Ohnishi J, Hirohara H, Hori K. Occurrence of highly yielded lectins homologous within the genus Eucheuma. J Appl Phycol. 1999;11:149–156. doi: 10.1023/A:1008062127564. [DOI] [Google Scholar]
- Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- Koharudin LMI, Gronenborn AM. Structural basis of the anti-HIV activity of the cyanobacterial Oscillatoria agardhii agglutinin. Structure. 2011;19:1170–1181. doi: 10.1016/j.str.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LMI, Gronenborn AM. Antiviral lectins as potential HIV microbicides. Curr Opin Virol. 2014;7:95–100. doi: 10.1016/j.coviro.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LMI, Furey W, Gronenborn AM. Novel fold and carbohydrate specificity of the potent anti-HIV cyanobacterial lectin from Oscillatoria agardhii. J Biol Chem. 2011;286:1588–1597. doi: 10.1074/jbc.M110.173278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LMI, Kollipara S, Aiken C, Gronenborn AM. Structural insights into the anti-HIV activity of the Oscillatoria agardhii agglutinin homolog lectin family. J Biol Chem. 2012;287:33796–33811. doi: 10.1074/jbc.M112.388579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJM. Nucleocytoplasmic plant lectins. Biochim Biophys Acta. 2010;1800:190–201. doi: 10.1016/j.bbagen.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJM. Lectin domains at the frontiers of plant defense. Front Plant Sci. 2014;5:397. doi: 10.3389/fpls.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, O’Keefe BR, Sowder RC, 2nd, Bringans S, Gardella R, Berg S, Cochran P, Turpin JA, Buckheit RW, Jr, McMahon JB, Boyd MR. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J Biol Chem. 2005;280:9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Cumsky MG, Zusman DR. Localization of myxobacterial hemagglutinin in the periplasmic space and on the cell surface of Myxococcus xanthus during developmental aggregation. J Biol Chem. 1981;256:12589–12595. [PubMed] [Google Scholar]
- O’Keefe BR, Smee DF, Turpin JA, Saucedo CJ, Gustafson KR, Mori T, Blakeslee D, Buckheit R, Boyd MR. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob Agents Chemother. 2003;47:2518–2525. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PK, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL, McCray PB., Jr Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol. 2010;84:2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno T, Shao H, Asada H, Shiraki K, Takahashi M, Yamanishi K. Analysis of human herpesvirus 6 glycoproteins recognized by monoclonal antibody OHV1. J Gen Virol. 1992;73:443–447. doi: 10.1099/0022-1317-73-2-443. [DOI] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HS. The culture of the red algal genus Eucheuma in the Philippines. Aquaculture. 1974;3:425–439. doi: 10.1016/0044-8486(74)90009-X. [DOI] [Google Scholar]
- Sato T, Hori K. Cloning, expression, and characterization of a novel anti-HIV lectin from the cultured cyanobacterium, Oschillatoria agardhii. Fish Sci. 2009;75:743–753. doi: 10.1007/s12562-009-0074-4. [DOI] [Google Scholar]
- Sato Y, Murakami M, Miyazawa K, Hori K. Purification and characterization of a novel lectin from a freshwater cyanobacterium, Oscillatoria agardhii. Comp Biochem Physiol B. 2000;125:169–177. doi: 10.1016/S0305-0491(99)00164-9. [DOI] [PubMed] [Google Scholar]
- Sato Y, Okuyama S, Hori K. Primary structure and carbohydrate binding specificity of a potent anti-HIV lectin isolated from the filamentous cyanobacterium Oscillatoria agardhii. J Biol Chem. 2007;282:11021–11029. doi: 10.1074/jbc.M701252200. [DOI] [PubMed] [Google Scholar]
- Sato Y, Morimoto K, Hirayama M, Hori K. High-mannose-specific lectin (KAA-2) from the red alga Kappaphycus alvarezii potently inhibits influenza virus infection in a strain-independent manner. Biochem Biophys Res Commun. 2011;405:291–296. doi: 10.1016/j.bbrc.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hirayama M, Morimoto K, Yamamoto N, Okuyama S, Hori K. High-mannose-binding lectin with preference for the cluster of α1-2-mannose from the green alga Boodlea coacta is a potent entry inhibitor of HIV-1 and influenza viuses. J Biol Chem. 2011;286:19446–19458. doi: 10.1074/jbc.M110.216655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Morimoto K, Kubo T, Yanagihara K, Seyama T. High-mannose-binding antiviral lectin PFL from Pseudomonas fluorescens Pf0-1 promotes cell death of gastric cancer cell MKN28 via interaction with α2-integrin. PLoS One. 2012;7:e45922. doi: 10.1371/journal.pone.0045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MD, Winter HC, Goldstein IJ, Markovitz DM. A lectin isolated from bananas is a potent inhibitor of HIV replication. J Biol Chem. 2010;285:8646–8655. doi: 10.1074/jbc.M109.034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove J, Fouquaert E, Smith DF, Proost P, Van Damme EJM. Lectin activity of the nucleocytoplasmic EUL protein from Arabidopsis thaliana. Biochem Biophys Res Commun. 2011;414:101–105. doi: 10.1016/j.bbrc.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Whitley MJ, Furey W, Kollipara S, Gronenborn AM. Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity. FEBS J. 2013;280:2056–2067. doi: 10.1111/febs.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvrouw M, Fikkert V, Hantson A, Pannecouque C, O’Keefe BR, McMahon J, Stamatatos L, de Clercq E, Bolmstedt A. Resistance of human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. J Virol. 2005;79:7777–7784. doi: 10.1128/JVI.79.12.7777-7784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Hoorelbeke B, Kagiampakis I, Demeler B, Balzarini J, LiWang PJ. The Griffithsin dimer is required for high-potency inhibition of HIV-1: evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob Agents Chemother. 2013;57:3976–3989. doi: 10.1128/AAC.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiie T, Maeda M, Kimura M, Hama Y, Uchida M, Kimura Y. Structural features of N-glycans of seaweed glycoproteins: Predominant occurrence of high-mannose type N-glycans in marine plants. Biosci Biotechnol Biochem. 2012;76:1996–1998. doi: 10.1271/bbb.120463. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- Ziolkowska NE, Wlodawer A. Structural studies of algal lectins with anti-HIV activity. Acta Biochim Pol. 2006;53:617–626. [PubMed] [Google Scholar]
- Ziolkowska NE, O’Keefe BR, Mori T, Zhu C, Giomarelli B, Vojdani F, Palmer KE, McMhon JB, Wlodawer A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure. 2006;7:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


