Abstract
Many antimicrobial peptides (AMPs) have multiple antimicrobial immunity effects. One such class of peptides is temporins. Temporins are the smallest (AMPs) found in nature and are highly active against gram-positive bacteria. Nowadays, there was a rapid increase in the availability of the 3D structure of proteins in PDB (protein data bank). The conserved residues and 3D structural conformations of temporins (AMPs) were still unknown. The present study explores the sequence analysis, alpha-helical structural conformations of temporins. The sequence of temporins was deracinated from APD3 database, the three-dimensional structure was constructed by homology modeling studies. The sequence analysis results show that the conserved residues among the peptide sequences, the maximum of the sequences are 70% alike to each other. The secondary structure prediction results revealed that 99% of temporin (AMPs) exhibited in alpha-helical form. The 3D structure speculated using RAMPAGE exposes the alpha-helical conformation in all temporins (AMPs). The phylogenetic analysis reveals the evolutionary relationships of temporins (AMPs), which are branched into seven clusters. As a result, we identified a list of potential temporin AMPs which docked to the antiviral protein (MERS-CoV), it shows good protein-peptide binding. This computational approach may serve as a good model for the rationale design of temporin based antibiotics.
Electronic supplementary material
The online version of this article (10.1007/s10989-019-09951-y) contains supplementary material, which is available to authorized users.
Keywords: Temporins, Alpha helical, Amino acid composition, Structure prediction, Phylogenetic tree, Protein-peptide docking
Introduction
Antimicrobial Peptides
Almost all multicellular organisms have AMPs to protect themselves from pathogenic microbes and acts as the first line of defence (Hancock and Chapple 1999). AMPs are an integral part of the body's natural immune system which exhibits not only antimicrobial, anticancer, antitumor activity and but also displays immunomodulating properties (Zasloff 2002). Majority of the mode of action studies performed using AMPs have cell membranes as their targets, and a variety of models have been proposed so far to explain the formation of membrane pores (Yeaman and Yount 2003). The disruptive membrane models such as “barrel-stave,” (Melo et al. 2009) “toroidal pore” (Hof et al. 2001) or “carpet models” (Steiner et al. 1988) provide a deeper insight into the interaction of amphipathic alpha-helical peptides with lipid bilayers, which are often referred as molecular electroporation (Matsuzaki 1999). The variations in cell membrane lipid composition of eukaryotic and prokaryotic organisms form as the primary targets for AMPs. The specificity of AMPs to the target cells relies upon the peptides favourable interaction with the microbial targets, which allow them to lyse a specific cell without affecting the host cells (Hancock et al. 2006). Net charge, amphipathicity, and hydrophobicity of AMPs also play a crucial role in the association of these peptides to target cellular membranes from exerting antimicrobial activity. Diverse antimicrobial peptides have been reported from various ecological components such as plants, mammals, insects, and invertebrates. Currently, more than 2838 AMPs have been deposit in the antimicrobial peptide database.
Temporins
The temporins are small (10 to 14 amino acid residues), linear (AMPs) with a net cationic charge and an amidated C-terminus. These peptide classes are collectively known as temporins and have shown the highest activity among AMPs studied till date, against both human erythrocytes and bacterial strains. The mode of action of AMPs against bacterial cells has been extensively identified and documented; their antiviral functions were not fully understood (Skalickova et al. 2015). Recent findings have suggested the efficacy of various classes of AMPs and their specific mode of action against viral pathogens and the potential use of these AMPs as an antiviral therapy (Marcocci et al. 2018; Ahmed et al. 2019).
Due to the membranolytic effects of temporins, the pathogens were difficult to develop resistance. Therefore, temporins are prospective aspirants for the design of imminent antiviral drugs with an innovative approach However, there is no structural information is available for the temporins family of AMPs. Therefore, we investigated the in silico structural information and protein-peptide interactions of temporins AMPs against MERS-CoV. These in silico findings are thought to help designing new types of peptide based antiviral drugs.
Middle East Respiratory Syndrome-Coronavirus (MERS-CoV)
In 2012, MERS-CoV was recognized in Saudi Arabia and belongs to the Coronaviridae family, mostly reported among the individuals of the Middle East (Milne-Price et al. 2014). MERS-CoV protein is a main target for therapeutic development to obstruct membrane fusion and viral entry (Du et al. 2017). Since, there is no effective therapeutics for Middle East Respiratory Syndrome and this research attempts to find using peptide therapeutics by establishing a basis for the protein-peptide interactions through molecular docking analysis for the antiviral protein MERS-CoV.
Materials and Methods
Dataset
Temporin family of (AMPs) was chosen for the present study. The amino acid sequence of (AMPs)belonging to the entire temporin family was retrieved from the APD3 antimicrobial database (https://aps.unmc.edu/AP/) (Wang et al. 2009, 2016; Wang and Wang 2004). A total of 121 antimicrobial peptides of temporin family from various organisms were collected from the database. The sequence alignment was performed with the aid of MUSCLE program.(Edgar 2004; Zimmermann et al. 2018). Followed by performing Multiple sequence alignment (MSA), the temporin family was clustered based on sequence similarity. MMseqs2 and CD-Hit were used (https://weizhongli-lab.org/cdhit_suite/cgi-bin/index.cgi) for clustering of peptides (Steinegger and Söding 2017; Huang et al. 2010a, b; Li and Godzik 2006). Among the clustered peptides the conserved residues were determined using Weblogo (Crooks et al. 2004). The secondary structure prediction for the entire temporin family of peptides was performed to achieve the homology modeling. Further, the biofilm active temporin peptides were also predicted using the dPABBs module (Sharma et al. 2016). The half-life of the peptides was identified using the HLP, a server for predicting half-life of peptides (Sharma et al. 2014).
Homology Modelling of Temporin AMPs
The structural similarity of the sequences was performed using BLASTp search and the templates with similar structures in the PDB database were retrieved. Among the chosen 121 (AMPs) of temporins, 120 peptides were 3D-modelled using Modeller software Version: 9.14 and PEP-FOLD for obtaining the three-dimensional structure of the peptides. The structure of one of the temporin peptide (PDB ID: 2MAA) is retrieved from protein databank structure (Sali and Blundell 1993; Fiser et al. 2000; Martí-Renom et al. 2000; Thevenet et al. 2012; Shen et al. 2014; Webb and Sali 2016). The modeled peptide structures were compared and evaluated with the standard peptide structure and validated using the Ramachandran plot at RAMPAGE server (Lovell et al. 2003).
Phylogenetic Tree Construction
A phylogenetic analysis based on the peptide sequence was carried out using the maximum-likelihood method with MEGA 6.0 (Tamura et al. 2013). The tree was evaluated using iTOL (interactive Tree of Life) package (Letunic and Bork 2016). The pairwise distance was calculated and interactively visualizes their data in the form of the heat map (Babicki et al. 2016).
Molecular Docking
To determine the molecular interaction of tem_a and high half-life (in seconds) of temporin peptides with the antiviral protein (MERS-CoV) using Cluspro server, online based protein–protein, protein-peptide docking algorithm (Kozakov et al. 2013, 2017; Vajda et al. 2017). MERS-CoV spike protein retrieved from protein data bank database (PDB ID: 5X59). The selective temporin peptides were uploaded to the Cluspro server for docking analysis and the hydrogen bond interactions between the MERS-CoV antiviral protein with temporins are visualized by Discovery studio visualizer software 2017(Dassault Systemes, BIOVIA Corp., San Diego, CA, USA).
Results and Discussions
Sequence and its Physicochemical Properties
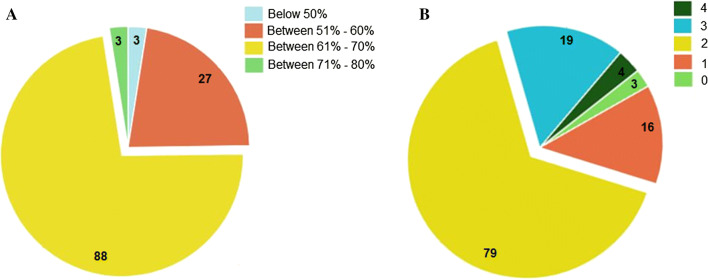
The amino acid sequence of temporin AMPs are listed from various Rana’s family, and these AMPs are showing high hydrophobicity and positive net charge (Fig. 1). Temporin AMPs sequence fasta file supplementary 1. The source and physicochemical properties of temporin AMPs were listed in Supplementary file 2. Most of the temporin AMPs are having more than 50% hydrophobicity (Fig. 1a), some of the temporin peptides (tem_hb1 ‘FLPLLAGLAAKWF’, tem_sh1 ‘FFFLSRIF’ and tem_1gd ‘FILPLIASFLSKFL’) has highest hydrophobicity percentage. In most cases, the hydrophobicity, length, net charges and other physicochemical parameters determined the antimicrobial activity of a peptide. In specific the hydrophobicity and charge of a peptide are critical factors determine the potency of an (AMPs) Any increase or decrease in the hydrophobicity index is directly proportional to the cationicity of a peptide. Most often the higher cationicity is attributing to greater binding efficiency, which results in effective binding of peptide over the bacterial surface (Malanovic and Lohner 2016). Once after binding the peptide takes its desired path to exert its action mechanism (Lee et al. 2002), while a decrease in hydrophobicity can reduce antimicrobial potency of the peptide (Kustanovich et al. 2002). During the synthesis of novel peptides, hydrophobicity is one of the perilous factors, for determining the range of target cells of AMP. Increasing the hydrophobicity of an AMP can alter the prospect of targets (Bahar and Ren 2013).
Fig. 1.
Pie chart depicting distribution of hydrophobicity and net charge of temporin AMPs
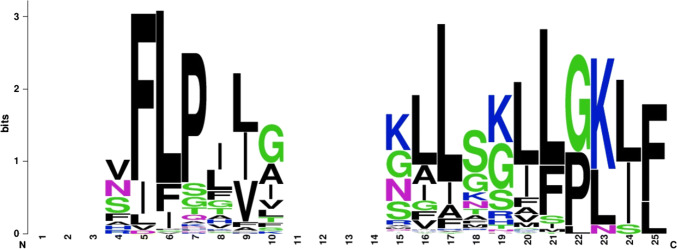
The net charge of all temporin AMPs contains a positive charge, and the average net charge is 2 (Fig. 1b). The selected AMPs of the net charge accounts for the ionizable groups of the peptide, resulting in the divergence from negative to positive and vice versa, which is the foremost constituent for the initial interaction with negatively charged cell membranes. The average length of the temporin AMPs is 13 residues. The length of an AMP is also a factor that attributes to the bioactivity of a peptide. A minimum of 7–8 amino acids are needed to form amphipathic structures with the hydrophobic and hydrophilic face on the opposite side of a peptide molecule (Zelezetsky and Tossi 2006). The overall height of the amino acids in (Fig. 2) indicates the conserved residues at that position. We identified that non-polar amino acids like phenylalanine, proline, leucine, shows the conserved among the temporin AMPs.
Fig. 2.
Conserved residues of temporin AMPs
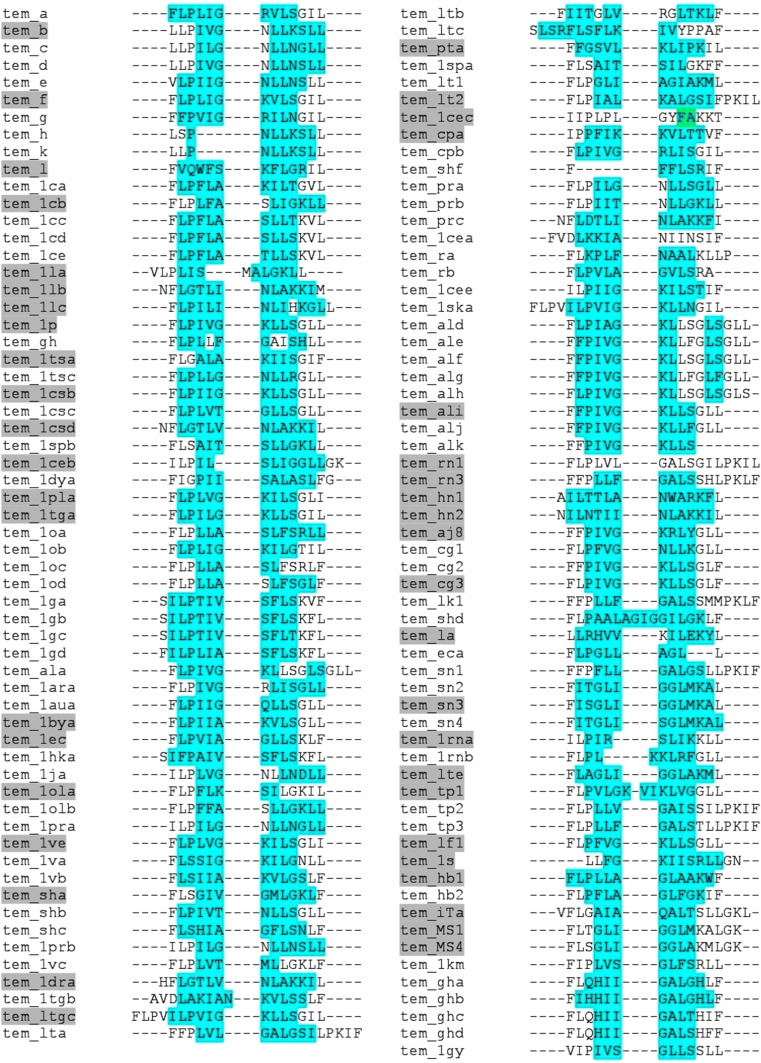
Sequence alignment of temporins AMPs represents the conserved residues among the peptide sequences, most of the temporin sequences are 70% similar to each other. The secondary structure prediction between the peptides shows the alpha helix and beta sheets, 98% of temporin AMPs exhibit alpha helical form. Some of the temporins display activity against the microbial biofilms, which are highlighted by grey colour shading (Fig. 3). From time to time, biofilms form a physical defense to protect bacteria from antibiotics.This might be due to slow development and reduced metabolic activity of a bacteria in such a population, the use of AMPs to inhibit biofilm formation could potentially be an efficient therapeutic option. In fact, due to the prevalent mechanism of action of AMPs, that depends on their ability to permeabilize or form pores within plasma membranes, they also have a strong capacity to act on increasing bacteria.(Batoni et al. 2011).A recent study on temporin anti-microbial peptides demonstrated significant anti-HSV-1 activity which was equivalent to that of Human cathelicidin LL-37 and a temporin synthetic analogue [K3]SHa. In addition, the study had emphasized its direct-acting mechanism for elimination of HSV-1(Roy et al. 2019). In an another in vitro anti-HSV-1 study using temporin B (TB), significant antiviral activity was observed which had resulted in a 5-log reduction of the viral titer. The disruption of the viral envelope was also confirmed upon observation using a transmission electron microscopy. Further, the TB was found to disturb the HSV-1 life cycle in different phases, including the attachment, entry into the host cell, and post-infection stage (Marcocci et al. 2018).
Fig. 3.
Multiple sequence alignment of temporin AMPs (Green colour indicates the sheet region; Magenta colour indicates the helix region; grey colour highlighted on temporins ID indicates the Bioactive peptides) (Color figure online)
Homology Modeling of Temporin AMPs and its Validation
Homology models of temporin AMPs have been constructed successfully for the identification of structural information. The result of all temporin peptides shows the α-helical conformation in modelled structures, which is an important class of AMPs holding higher amphipathicity. Moreover, the α-helical conformation allows effective interaction with biological membranes (Huang et al. 2010a, b). The modelled 3D structures of 120 temporins (AMPs) evaluated using RAMPAGE helps to interpret the allowing and disallowing regions, which declares the evaluation of peptide structures in Supplementary file 3.
Evolutionary Relationship and Recognition of Half-Life of Temporin AMPs
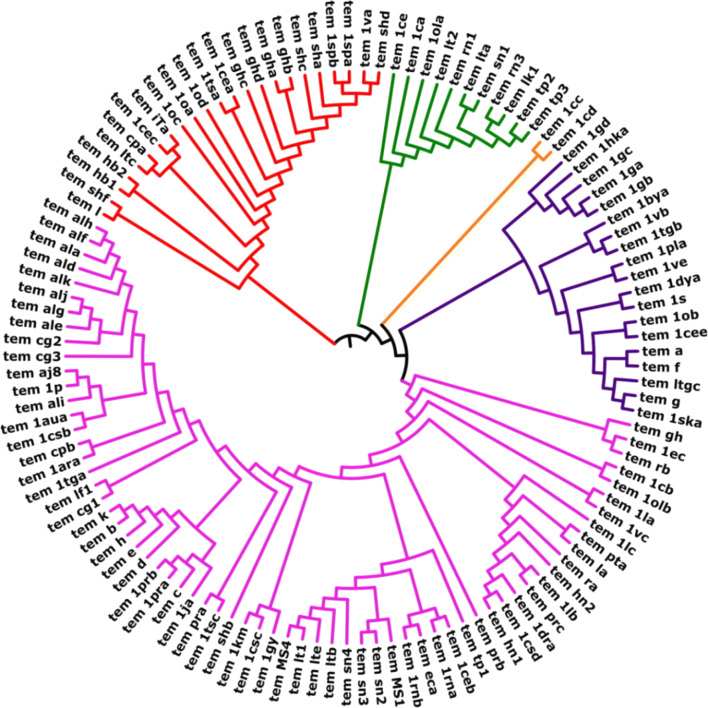
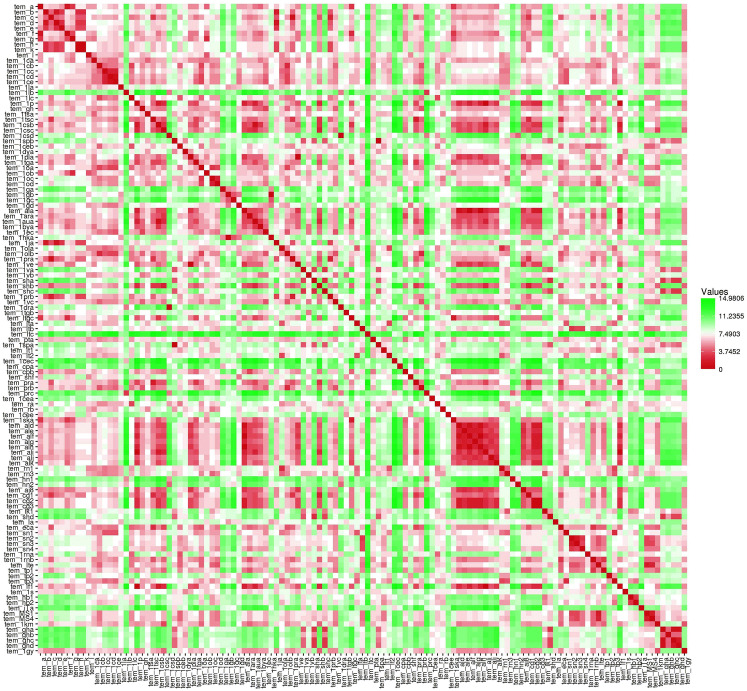
We have constructed a phylogeny tree of all the 121 temporin AMPs (Fig. 4) to evaluate the evolutionary relationship between sequences. Maximum likelihood approaches are used to evaluate the Phylogenetic trees for a set of species (Cho 2012), and it's a robust statistical foundation that allows comparison of different trees, parameters and models. Based on the phylogenetic tree (Fig. 4), the temporin peptide sequences classified into five clades (CLADE A, CLADE B, CLADE C, CLADE D, and CLADE E). Each clade node represents the individual colour to which the branched group has been recognized (Fig. 4). The Pairwise distance matrix was calculated for all temporin peptides from MEGA software (Supplementary file 4) and a heatmap was constructed using HEATMAPPER, which shows the graphical representation of sequence pairwise between the temporin AMPs (Fig. 5).
Fig. 4.
The phylogenetic tree of temporin AMPs
Fig. 5.
A heat map showing the sequence identity between the temporin AMPs (red colour indicates the most similar pairwise distance green colour indicates the least similiar pairwise distance) (Color figure online)
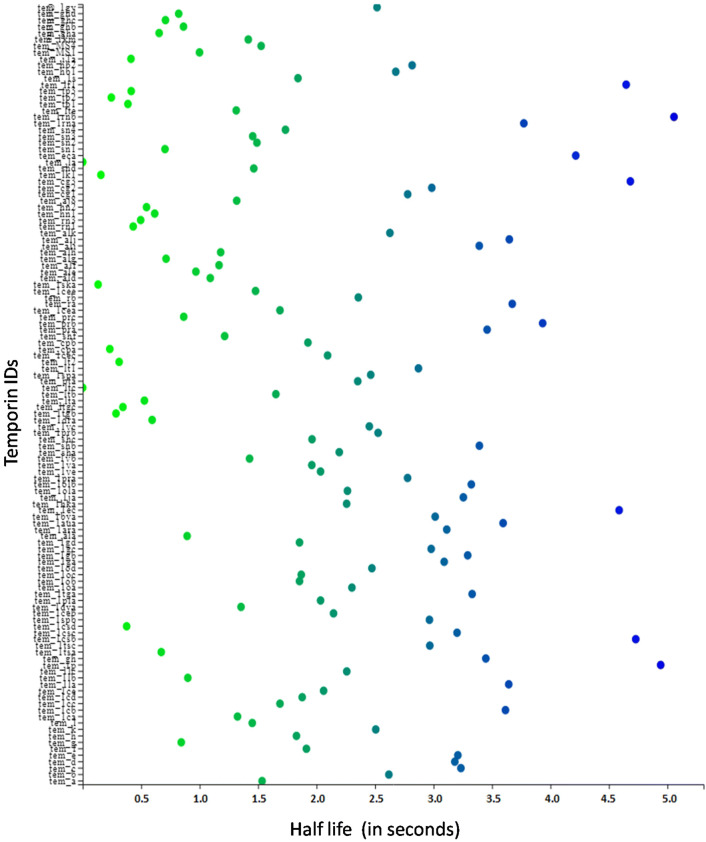
Plenty of the (AMPs) have entered clinical trials, but not many have been supported by the U.S. Food and Drug Administration to date due to the reason of short half-life. In this work, we predicted the short half-life of temporin AMPs by insilico method. (Fig. 6) shows the scatter visualization of predicted half-life values of temporin AMPs. Peptides like tem_1rnb, tem_1p, tem_1csb, tem_cg3, tem_lf1, tem_1ec, tem_eca are showing the higher half-life values in seconds. The lists of the half-life of all temporin AMPs Supplementary file 4.
Fig. 6.
A scatter plot showing the half-life of the temporin AMPs (Green colour represent the minimum half-life, canal blue colour represent the neutral half-life, navy colour represent the maximum half-life) (Color figure online)
Evaluation of MERS CoV – Temporin AMPs and its Interaction Analysis
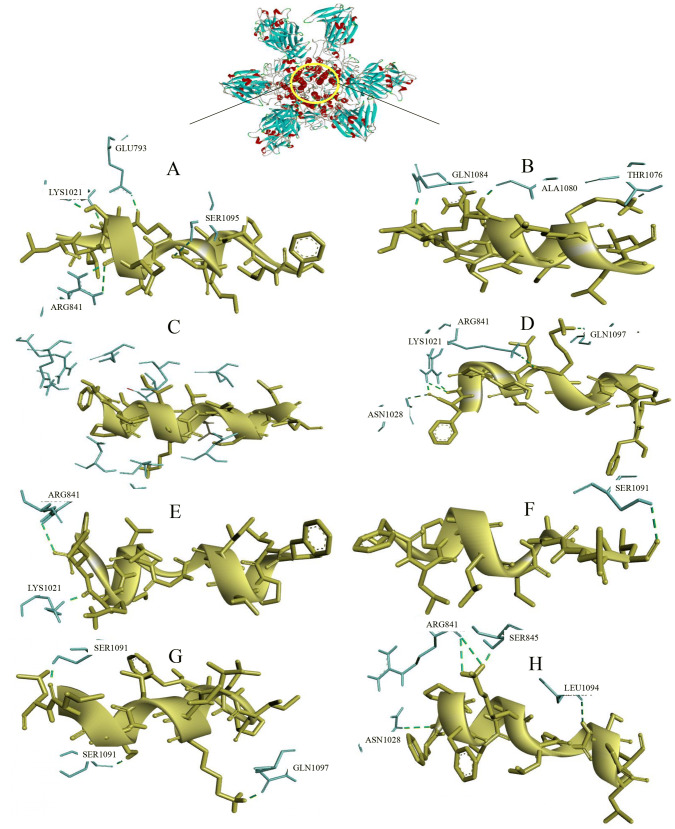
The molecular docking of selective temporin AMPs with antiviral protein (MERS-CoV) was performed by using cluspro online server. For further evauation, we selected eight antimicrobial peptides, namely, tem_a, tem_1rnb, tem_1p, tem_1csb, tem_cg3, tem_lf1, tem_1ec and tem_eca belonging to family of temporins. The modeled, validated structure of tem_1rnb, tem_1p, tem_1csb, tem_cg3, tem_lf1, tem_1ec and tem_ecahave been chosen as ligand for this protein-peptide docking studies. The docked pose with the best fit was selected for each peptide-protein complex based on same binding region (Fig. 7). The cluspro result of MERS CoV – Temporin AMPs complex details presented in Table 1.
Fig. 7.
Visualization of docked complexes (tem_a—MERS-CoV complex Fig. 5a, tem_1rnb—MERS-CoV Fig. 5b, tem_1csb—MERS-CoV complex Fig. 5c, tem_cg3—MERS-CoV complex Fig. 5d, tem_1ec—MERS-CoV complex Fig. 5e, tem_eca—MERS-CoV complex Fig. 5f, tem_1p—MERS-CoV complex Fig. 5g, tem_lf1—MERS-CoV complex Fig. 5h)
Table 1.
Energy of the docked complex based on cluster size (member), energy scores
| Peptide | Target | Representative | Member | Energy score |
|---|---|---|---|---|
| Tem_a | MERs-CoV | Center | 277 | − 874.5 |
| Tem_1rnb | 115 | − 789.6 | ||
| Tem_1p | 223 | − 925.0 | ||
| Tem_1csb | 133 | − 890.5 | ||
| Tem_cg3 | 133 | − 861.0 | ||
| Tem_1ec | 191 | − 1000.8 | ||
| Tem_1f1 | 133 | − 882.1 | ||
| Tem_eca | 917 | − 784.9 |
Further, the protein-peptide complexes were visualized by Discovery studio visualizer, to display hydrogen bond interaction involved in the protein-peptide docking. The tem_a peptide shows good binding with the MERS-CoV, it forms six hydrogen bond interaction with GLU793, SER1095, LYS1021, ARG841 (Fig. 5a); tem_1rnb forms hydrogen bond interaction with GLN1084, ALA1080, THR1076 (Fig. 5b); tem_1csb shows null hydrogen bond interaction with the target (Fig. 5c); tem_cg3 forms six hydrogen bond interaction with ARG841, GLN1097, LYS1021, ASN1028 (Fig. 5d); tem_1ec shows 2 hydrogen bond interaction with ARG841, LYS1021(Fig. 5E); tem_eca forms one hydrogen bond with SER1091 (Fig. 5f); tem_1p forms three hydrogen bond interaction with SER1091, GLN1097 (Fig. 5g); tem_lf1 shows good interactions, it forms five hydrogen bonds with LEU1094, SER845, ARG841, ASN1028 (Fig. 5h). Recently, Mustafa et al. 2019 had docked, investigated and validated the interactions between the various peptides (Cyclotides, Chicken beta defensin, Dermaseptin-s9, Human alpha defensin, Plectasin, Rat defensin, Rat alpha defensin) against MERS-CoV and has demonstrated prominant antiviral activity.
Conclusion
In this present study, we have interpreted the sequence, modelled 3D structures, evolutionary relationship and docking studies with antiviral protein (MERS-CoV) of temporin AMPs through a homology modeling, phylogeny tree construction and protein peptide docking. The 3D structural of temporin antimicrobial peptides successfully predicted and results shows the alpha-helical structural diversity among the peptides. Phylogenetic analysis exposes that all temporin peptides are 70% evolutionary similar to each other. Some of the temporin peptides like tem_a and high scored half-life temporin peptides were docked to MERS-CoV protein, and results shows good interaction with the antiviral protein. These observations reinforce the idea that temporin AMPs may serve as a new therapeutic application for the future development. Furthermore, the findings reported above are preliminary and will certainly require experimental evaluation through in vitro and in vivo studies, which are crucial for their development into therapeutic targets against MERS-CoV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
A special thanks to Department of Pharmaceutical Technology, University College of Engineering, Bharathidasan Institute of Technology Campus, Anna University, Tiruchirappalli-620024 and Center for Research, Anna University, Chennai-600025, Tamil Nadu, for support and facilities provided.
Compliance with Ethical Standards
Conflict of interest
Authors declare no conflict of interest.
Research Involving Human and Animal Participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmed A, Siman-Tov G, Hall G, et al. Human antimicrobial peptides as therapeutics for viral infections. Viruses. 2019;11:704. doi: 10.3390/v11080704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoni G, Maisetta G, Brancatisano FL, et al. Use of antimicrobial peptides against microbial biofilms: advantages and limits. Curr Med Chem. 2011;18:256–279. doi: 10.2174/092986711794088399. [DOI] [PubMed] [Google Scholar]
- Cho A (2012) Constructing phylogenetic trees using maximum likelihood. Scripps Sr Theses
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Yang Y, Zhou Y, et al. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Do RKG, Šali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Chapple DS. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/AAC.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock REW, Brown KL, Mookherjee N. Host defence peptides from invertebrates—emerging antimicrobial strategies. Immunobiology. 2006;211:315–322. doi: 10.1016/j.imbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- van t Hof W, Veerman ECI, Helmerhorst EJ, Amerongen AVN. Antimicrobial peptides: properties and applicability. Biol Chem. 2001;382:597–619. doi: 10.1515/BC.2001.072. [DOI] [PubMed] [Google Scholar]
- Huang Y, Huang J, Chen Y. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell. 2010;1:143–152. doi: 10.1007/s13238-010-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, et al. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Beglov D, Bohnuud T, et al. How good is automated protein docking? Proteins Struct Funct Bioinforma. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustanovich I, Shalev DE, Mikhlin M, et al. Structural requirements for potent versus selective cytotoxicity for antimicrobial dermaseptin S4 derivatives. J Biol Chem. 2002;277:16941–16951. doi: 10.1074/jbc.M111071200. [DOI] [PubMed] [Google Scholar]
- Lee DG, Kim HN, Park Y, et al. Design of novel analogue peptides with potent antibiotic activity based on the antimicrobial peptide, HP (2–20), derived from N-terminus of Helicobacter pylori ribosomal protein L1. Biochim Biophys Acta. 2002;1598:185–194. doi: 10.1016/S0167-4838(02)00373-4. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct Funct Bioinforma. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Malanovic N, Lohner K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals. 2016;9:59. doi: 10.3390/ph9030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcocci ME, Amatore D, Villa S, et al. The amphibian antimicrobial peptide temporin b inhibits in vitro herpes simplex virus 1 infection. Antimicrob Agents Chemother. 2018;62:e02367. doi: 10.1128/AAC.02367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí-Renom MA, Stuart AC, Fiser A, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462:1–10. doi: 10.1016/S0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- Melo MN, Ferre R, Castanho MARB. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat Rev Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- Milne-Price S, Miazgowicz KL, Munster VJ. The emergence of the middle east respiratory syndrome coronavirus. Pathog Dis. 2014;71:121–136. doi: 10.1111/2049-632X.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa S, Balkhy H, Gabere M. Peptide-protein interaction studies of antimicrobial peptides targeting middle east respiratory syndrome coronavirus spike protein: an in silico approach. Adv Bioinform. 2019;2019:1–16. doi: 10.1155/2019/6815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, Lebeau L, Chessa C, et al. Comparison of anti-viral activity of frog skin anti-microbial peptides temporin-sha and [K3]SHa to LL-37 and temporin-Tb against herpes simplex virus type 1. Viruses. 2019;11:77. doi: 10.3390/v11010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sharma A, Singla D, Rashid M, Raghava GP. Designing of peptides with desired half-life in intestine-like environment. BMC bioinform. 2014;15(1):282. doi: 10.1186/1471-2105-15-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Gupta P, Kumar R, Bhardwaj A. dPABBs: a novel in silico approach for predicting and designing anti-biofilm peptides. Sci Rep. 2016;6:21839. doi: 10.1038/srep21839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J Chem Theory Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- Skalickova S, Heger Z, Krejcova L, et al. Perspective of use of antiviral peptides against influenza virus. Viruses. 2015;7:5428–5442. doi: 10.3390/v7102883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M, Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017 doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- Steiner H, Andreu D, Merrifield RB. Binding and action of cecropin and cecropin analogues: antibacterial peptides from insects. Biochim Biophys Acta. 1988;939:260–266. doi: 10.1016/0005-2736(88)90069-7. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet P, Shen Y, Maupetit J, et al. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S, Yueh C, Beglov D, et al. New additions to the ClusPro server motivated by CAPRI. Proteins Struct Funct Bioinform. 2017;85:435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Res. 2004;32:590D–592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2016;86:2.9.1–2.9.37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zelezetsky I, Tossi A. Alpha-helical antimicrobial peptides—using a sequence template to guide structure—activity relationship studies. Biochim Biophys Acta. 2006;1758:1436–1449. doi: 10.1016/j.bbamem.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam S-Z, et al. A completely reimplemented MPI bioinformatics toolkit with a new hhpred server at its core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/J.JMB.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.