Abstract
This study aimed to determine the aetiology of community-acquired pneumonia (CAP) by adding polymerase chain reaction (PCR) to conventional methods and to describe the clinical and laboratory features between patients with bacterial pneumonia (BP) and viral pneumonia (VP). Adults with CAP admitted from November 2009 to October 2010 were included. Demographics, comorbidities, severity and clinical features were recorded. Conventional microbiological methods included blood and sputum cultures, acute and convalescent serologic samples, and antigen urinary detection. New methods included multiplex PCR for Mycoplasma pneumoniae, Legionella pneumophila, Chlamydophila pneumoniae, Bordetella pertussis and 15 respiratory viruses. A total of 169 patients were included. Using conventional methods, we identified a pathogen in 51 % of cases. With PCR, up to 70 % of cases had an aetiological diagnosis. Forty-five patients had BP (34 %), 22 had VP (17 %) and 25 (19 %) had co-infection (BP and VP). Pneumococci and respiratory syncytial virus (RSV) were the most frequently identified pathogens. Procalcitonin (PCT) and C-reactive protein (CRP) median values were significantly higher in BP than in VP patients. Shaking chills, higher CURB score and shock were significantly more frequent in BP. A viral infection was identified in more than one-third of patients with CAP. Clinical and laboratory features could help to differentiate between VP and BP and to guide empirical therapy.
Keywords: Influenza, Respiratory Syncytial Virus, Severe Acute Respiratory Syndrome, Multiplex Polymerase Chain Reaction, Respiratory Virus
Introduction
Community-acquired pneumonia (CAP) is a common disease, representing the most frequent cause of hospital admission and mortality of infectious origin in developed countries [1]. Until recently, despite the availability of many diagnostic techniques, nearly half of the cases of CAP remained undiagnosed [2], representing a risk for inappropriate antimicrobial therapy.
Etiologic investigations in CAP have traditionally focused on bacterial agents and little attention has been paid to viral causes. A number of reasons have been brought forward to explain this; first, it is still unclear whether a virus by itself can cause pneumonia or whether the virus acts in conjunction with other respiratory pathogens; second, diagnosis is difficult, primarily because the methods used up until recently (culture, immunofluorescence for viral antigens and serology) are expensive and often unavailable; and third, because of the absence of specific antiviral agents.
In adults, influenza remains the predominant viral cause of CAP; however, the Infectious Diseases Society of America/American Thoracic Society (IDSA/ATS) guidelines for CAP include other commonly recognised viruses, such as respiratory syncytial virus (RSV), adenovirus and parainfluenza virus, as well as less common viruses, including human metapneumovirus, herpes simplex virus, varicella-zoster virus, severe acute respiratory syndrome (SARS)-associated coronavirus and measles virus [3].
Recently, polymerase chain reaction (PCR)-based methods have been developed for the diagnosis of many bacterial and a large number of viral respiratory pathogens [4].
In this study, we aimed to describe the aetiology of CAP in patients admitted to hospital and to describe the clinical and laboratory features of patients with bacterial pneumonia (BP) compared to those with viral pneumonia (VP) in an H1N1 pandemic year.
Materials and methods
Setting and study design
This prospective, observational study was conducted at Hospital Universitari Mutua Terrassa, a 500-bed acute care hospital with approximately 26,000 admissions/year for a population of 350,000 inhabitants.
Inclusion criteria and clinical variables
From November 2009 to October 2010, consecutive adults with CAP admitted to hospital for at least 24 h from Monday to Friday were included. Only patients with a good quality respiratory sample for microbiological analysis were included. CAP was defined as an acute illness with radiographic pulmonary shadowing confirmed by one clinician and one radiologist and at least two of the following: fever, cough, dyspnoea, sputum production, pleuritic chest pain or shaking chills. Exclusion criteria were: age <18 years, pneumonia distal to endobronchial obstruction, pulmonary tuberculosis, bronchiectasis and nosocomial pneumonia.
On admission, data were prospectively collected and included demographic characteristics, smoking, co-morbidities, Charlson score [5] and prognosis measured by the pneumonia severity index (PSI) [6] and the CURB score (confusion, urea >7 mmol/L, respiratory rate >30 breaths per/min and blood systolic pressure <90 mm Hg or diastolic <60 mm Hg) [7], immunosuppressive conditions, clinical features, biomarkers [C-reactive protein (CRP) and procalcitonin (PCT)], empirical and definitive antimicrobial therapy, length of stay and in-hospital mortality. We defined immunosuppression as patients on current chemotherapy, biological therapy, chronic systemic steroid use (15 mg/d for 14 days or equivalent dose), neutropaenia or HIV with CD4 count less than 200 cells/mm3.
The study was conducted in accordance with ethical principles set out in the latest version of the Declaration of Helsinki and the standard used for Good Clinical Practice, and was approved by the hospital Ethics and Research Committee. Written informed consent was obtained from all patients.
Microbiological studies
At baseline, two sets of blood cultures were obtained in all patients prior to commencing antibiotic therapy. Blood cultures were processed with the system BacT-Alert® (bioMérieux, Marcy-Etoile, France). Nasopharyngeal swab specimens for the diagnosis of influenza A pandemic H1N1 strain were collected during the pandemic. Sputum samples were processed for Gram stain. Only samples containing a predominance of polymorphonuclear leukocytes and a few squamous epithelial cells were considered to be acceptable for culture. Viral RNA and DNA were extracted from each sputum sample using the kit Seeplex RV15 ACE Detection (Seegene®), following the manufacturer’s instructions.
PCR was performed in sputum samples for Legionella pneumophila, Mycoplasma pneumoniae, Chlamydophila pneumoniae, Bordetella pertussis and multiplex PCR for respiratory viruses (detection by capillary electrophoresis, Seegene®); influenza A and B, RSV A and B, parainfluenza 1, 2, 3 and 4, coronavirus 229/NL63E, OC43/HKU1, rhinovirus A and B, adenovirus, metapneumovirus, bocavirus 1, 2, 3 and 4, and enterovirus.
The aetiology was also pursued by searching for Streptococcus pneumoniae and L. pneumophila serogroup I antigens in urine (BinaxNOW® Urinary Antigen Test; Binax).
Acute and convalescent serum samples 3 to 6 weeks later were run in parallel for the serologic diagnosis of L. pneumophila, M. pneumoniae, C. pneumoniae and Coxiella burnetii. The serology was considered to be positive when the IgM titre was ≥1/10, when there was a seroconversion of IgG (shown in acute and convalescent at 4 weeks) and were available if only a single sample of serum positivity was assessed when the IgG was >1/256.
Conventional microbiological diagnostic methods included two sets of blood cultures, sputum stain and culture, and the detection of S. pneumoniae or L. pneumophila antigens in urine. In all the patients, all methods were used. Cases in which a virus and bacteria were simultaneously identified were defined as co-infection. When two viruses were identified, CAP was defined as dual VP. In order to compare the clinical and laboratory features between BP versus VP, patients with co-infection were excluded.
Only the diagnostic tests at admission were protocolised. After that, the physicians in charge of the included patients were responsible for performing appropriate diagnostic tests based on epidemiology, clinical characteristics and the patient’s clinical course.
Statistical analysis
Values are presented as frequencies (%) for qualitative variables and mean ± standard deviation (SD) for continuous, normally distributed variables or median (interquartile range) for those not having a normal distribution. Comparisons between patients with the diagnosis of BP or VP were performed by using the unpaired t-test or Mann–Whitney U-test when appropriate. Chi-square was used for comparing proportions between these two groups. Statistical significance was taken as a p-value ≤ 0.05. Data analyses were performed with the use of SPSS software, version 13.
Results
A total of 169 patients were enrolled. Thirty-eight (22 %) patients without sputum sample (lack of production or missed collection) were excluded. Thus, a total of 131 patients constituted the final evaluable population.
In 48.4 % of patients, the sample was thought to be good for bacterial culture (predominance of leukocytes and the presence of few squamous epithelial cells). Acute serum samples were lost or not done in 13 patients (10 %) and convalescent serum samples for serology were not obtained in 44 patients (34 %) of the study group. Causes for the lack of a second sample were loss of follow-up or death (seven patients).
An aetiological diagnosis using conventional diagnostic methods could be established in 66/131 (51 %) patients. When real-time PCR was included, at least one aetiologic agent was identified in 92/131 (70 %) patients.
Aetiological diagnosis
According to the aetiological diagnosis, 45 (34 %) cases of CAP were due to bacteria, 22 (17 %) were due to viruses, 25 (19 %) were due to viruses and bacteria (co-infection), and 39 (30 %) were of unknown aetiology.
Figure 1 shows the most common pathogens in patients diagnosed as having BP. S. pneumoniae was identified in 28 patients. The remaining bacterial agents were responsible for less than five cases. In four patients, pneumococcal infection was associated with other bacterial agents (C. burnetii in two, M. pneumoniae in one and C. pneumoniae in one patient).
Fig. 1.
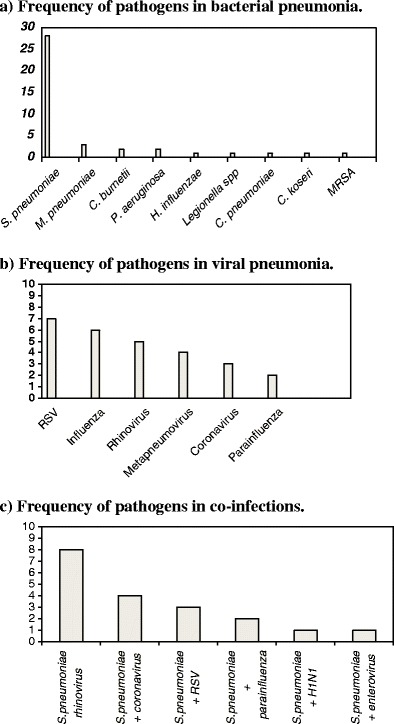
a The frequency of pathogens in bacterial pneumonia (BP). b The frequency of pathogens in viral pneumonia (VP). c The frequency of pathogens in co-infections
The contribution of the different diagnostic methods for the diagnosis of BP is illustrated in Table 1. Blood cultures provided a microbiological diagnosis for 16 (12 %) of 131 patients, 15 S. pneumoniae and one Pseudomonas aeruginosa in a patient receiving immunosuppressive therapy. S. pneumoniae was detected by a positive urinary antigen assay in 42 out of the 131 patients tested.
Table 1.
Microbiological diagnosis of patients with bacterial pneumonia (BP)
| Aga, n | Sputum culture, n | BCb, n | PCR multiplex, n | Telescoped brush, n | Serology, n | |
|---|---|---|---|---|---|---|
| L. pneumophila, n = 1 | 1 | Negative | Negative | Negative | Not done | Negative |
| S. pneumoniae, n = 51 | 42 |
Gram 16 Culture 14 |
15 | Negative | Not done | Not appropriate |
| P. aeruginosa, n = 3 | Not appropriate | Culture 2 | 1 | Negative | Not done | Not appropriate |
| M. pneumoniae, n = 6 | Not appropriate | Negative | Negative | 3 | Not done | 3 |
| C. pneumoniae, n = 3 | Not appropriate | Negative | Negative | 3 | Not done | Negative |
| C. burnetii, n = 4 | Not appropriate | Negative | Negative | Negative | Not done | 4 |
| Citrobacter koseri, n = 1 | Not appropriate |
Gram 1 Culture 1 |
Negative | Negative | Not done | Not appropriate |
| Escherichia coli, n = 1 | Not appropriate | Negative | Negative | Negative | 1 | Not appropriate |
| MRSAc, n = 1 | Not appropriate | Negative | Negative | Negative | 1 | Not appropriate |
| H. influenzae, n = 2 | Not appropriate | Culture 2 | Negative | Negative | Not done | Not appropriate |
a Ag urine antigen
b BC blood culture
c MRSA methicillin-resistant Staphylococcus aureus
Among the 22 patients with VP, the most common organisms were RSV (n = 7) and influenza A and B (n = 6, two of them H1N1). Other aetiological agents included parainfluenza, rhinovirus, metapneumovirus and coronavirus (Fig. 1). There were four dual infections of RSV with other viruses (two parainfluenza, one rhinovirus and one coronavirus).
In patients with co-infection, the most common combinations were S. pneumoniae and rhinovirus (n = 8), followed by S. pneumoniae and coronavirus (n = 4), and S. pneumoniae and RSV (n = 3). Figure 1 summarises the most frequent co-infections.
Our design allowed us to learn something about the seasonality of the respiratory viruses studied. Influenza and RSV were the two most frequent viruses in late autumn and early winter, followed by coronavirus and parainfluenza. In spring and summer, only metapneumovirus was identified. Rhinovirus was found throughout the year (Fig. 2).
Fig. 2.
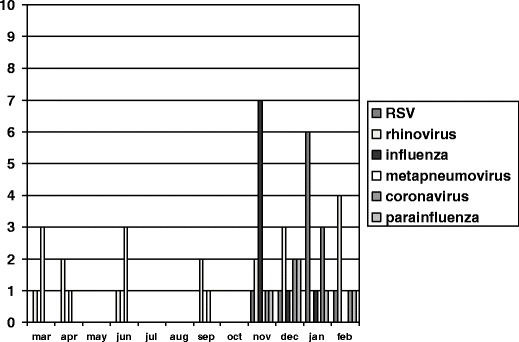
Seasonality of respiratory virus infections in 2009 and 2010
Viral pneumonia versus bacterial pneumonia
Comparing VP and BP, no differences were found in terms of age, gender or co-morbidities in the Charlson score. The proportion of smokers was higher in the group with VP (see Table 2).
Table 2.
Demographics
| VP, n = 22 | BP, n = 45 | p-value (VP and BP) | Unknown, n = 39 | Mixed infection, n = 25 | |
|---|---|---|---|---|---|
| Age, years [range] | 56 [48–64] | 60 [54–67] | NSa | 68 [61–75] | 74 [68–80] |
| Male sex | 17 (77 %) | 32 (71 %) | NSa | 24 (61 %) | 17 (68 %) |
| Current smoker | 9/14 (64 %) | 11/36 (30 %) | 0.05 | 13/28 (46 %) | 8/47 (47 %) |
| Immunosuppression, n = 12 | 2 (9 %) | 5 (11 %) | NSa | 3 (8 %) | 2 (8 %) |
| Charlson score | 1.4 [0.6-2.3] | 1.9 [1.2-2.6] | NSa | 1.7 [1–2.3] | 2.2 [1.3-3.1] |
aNot significant
VP was characterised by a lower frequency of shaking chills and higher frequencies of myalgia, rhinorrhoea or odynophagia and dry cough (see Table 3).
Table 3.
Clinical characteristics of bacterial pneumonia (BP) versus viral pneumonia (VP)
| VP, n = 22 | BP, n = 45 | p-value (VP and BP) | Unknown, n = 39 | Mixed infection, n = 25 | |
|---|---|---|---|---|---|
| Hours with symptoms [range] | 127 [31–223] | 99 [57–140] | NSa | 155 [90–221] | 86 [34–138] |
| Pleurisy pain | 6 (27 %) | 20 (44 %) | NSa | 13 (33 %) | 7 (28 %) |
| Shaking chills | 1 (4 %) | 21 (47 %) | <0.001 | 8 (20 %) | 4 (16 %) |
| Mental confusion | 1 (4 %) | 3 (7 %) | NSa | 0 | 1 (4 %) |
| Myalgia or rhinorrhoea or odynophagia | 10 (45 %) | 9 (20 %) | 0.04 | 8 (20 %) | 5 (20 %) |
| Headache | 1 (4 %) | 1 (2 %) | NSa | 2 (5 %) | 1 (4 %) |
| Dry cough | 10 (45 %) | 9 (20 %) | 0.04 | 5 (13 %) | 5 (20 %) |
| Productive sputum | 11 (50 %) | 27 (60 %) | NSa | 28 (71 %) | 16 (64 %) |
aNot significant
We found no differences between VP and BP on chest radiograph characteristics. Twenty (87 %) patients with VP presented with an alveolar consolidation.
In terms of laboratory findings, patients with VP were far more likely to have a low PCT level than those with BP. Also, the CRP level was lower (see Table 4).
Table 4.
Laboratory features in VP versus BP
| VP, n = 22 | BP, n = 45 | p-value (VP and BP) | Unknown, n = 39 | Mixed infection, n = 25 | |
|---|---|---|---|---|---|
| CRP (mg/L)a [range] | 145 [93–197] | 231 [181–281] | 0.051 | 190 [143–238] | 197 [126–269] |
| PCT (mg/L)b [range] | 0.8 [0.008-1.6] | 10 [3–18] | 0.03 | 3 [0.09–5.8] | 6 [1.3–9.7] |
| Creatinine [range] | 1.15 [0.8-1.4] | 1.18 [1–1.3] | NSc | 1.18 [0.9–1.4] | 2.4 [0.2–5] |
| CPK [range] | 67 [41–93] | 85 [45–126] | NSc | 99 [53–145] | 88 [44–131] |
| LDH [range] | 530 [380–508] | 444 [354–535] | NSc | 454 [370–538] | 419 [330–509] |
| Abnormal WBC >12 or <4 cells/mL | 11,227 [7,723–14,731] | 14,152 [11,472–16,833] | NSc | 11,522 [10,010–13,034] | 13,285 [10,477–16,094] |
a CRP C-reactive protein
b PCT procalcitonin
cNot significant
In terms of severity, the CURB score was higher in BP than in VP, and shock was only present in BP. The Charlson score, PSI, need of mechanical ventilation, intensive care unit (ICU) admission and mortality were similar between both groups (see Table 5). The global mortality rate was 5.3 % (7/131), being highest in patients with co-infection (12 %; 3/25).
Table 5.
Severity of disease. Differences in VP versus BP
| VP, n = 22 | BP, n = 45 | p-value (VP and BP) | Unknown, n = 39 | Mixed infection, n = 25 | |
|---|---|---|---|---|---|
| PSIa | 3 [2–3] | 3 [2–3] | NSc | 3 [3–4] | 4 [3–4] |
| CURB-65b | 0.7 [0.3–1.8] | 1.4 [1.4–1.7] | 0.02 | 1.2 [0.9–1.4] | 1.7 [1.2–2.2] |
| Shock | 0 | 8 (18 %) | 0.04 | 3 (8 %) | 1 (4 %) |
| Empyema | 0 | 5 (11 %) | NSc | 1 (3 %) | 0 |
| Death | 0 | 2(4 %) | NSc | 2 (5 %) | 3 (12 %) |
| Mechanical ventilation | 1 (5 %) | 2 (4 %) | NSc | 3 (8 %) | 1 (4 %) |
| ICU admission | 3 (15 %) | 8 (18 %) | NSc | 6 (15 %) | 1 (4 %) |
aPneumonia Severity Index
bCURB-65 score (confusion, urea >7 mmol/L, respiratory rate >30 breaths per/min and blood systolic pressure <90 mm Hg or diastolic <60 mm Hg, age >65 years)
cNot significant
Immunocompromised patients
Only 12 patients with immunosuppression were enrolled. Two of them were due to HIV infection with a CD4 count less than 200 cells/mm3, one was on corticosteroids treatment and one was on treatment with IFN beta, and, finally, eight patients were currently on antineoplastic agents. At follow-up, immunosuppressed patients received the following diagnoses: two VP (one was an HIV-infected patient who also presented an immune reconstitution inflammatory syndrome), five BP, two with mixed infection and three with unknown etiologies. In both HIV-infected patients, a fibrobronchoscopy and Gömöri trichome stain were done. Two immunosuppressed oncologic patients died due to severe sepsis with bacteraemia due to pneumococcal infection on the first day of admission.
Discussion
Establishing a microbiological diagnosis for patients with CAP is challenging. Although many studies have been performed, the aetiology remains unknown in approximately one-half of the cases [2]. In the present study, the use of PCR testing added to conventional microbiologic methods was able to produce 70 % of etiologic diagnoses, which represented an increase of 40 % in the number of diagnoses as compared with standard methods.
A quarter of our patients had a virus as the only pathogen isolated. Recent studies that have also sought to determine the presence of respiratory virus in CAP in adults with extensive diagnostic testing have reported an incidence of VP ranging from 5.6 to 50 % [8–19]. In a recently published large cohort, a similar rate of co-infection has been reported. However, we found a smaller proportion of dual viral infection as compared with that study focused on influenza-associated CAP [20, 21].
The role of respiratory viruses as causative agents of pneumonia is controversial. Lieberman et al. [18] used nucleic acid amplification techniques to provide data on the frequency and identification of viral causes in hospitalised patients with lower respiratory tract infections. They compared the findings in those with radiologically confirmed CAP to an ambulatory control population without respiratory tract infection and to a hospitalised population with non-pneumonic lower respiratory tract infection (NPLRTI). The frequency of viral CAP was 31.7 % of 183 patients, compared with 7.1 % of 450 controls and 51.7 % of 201 patients with NPLRTI. The frequency of viral CAP was put into context by demonstrating that viruses were even more common in patients with NPLRTIs and that the frequency of a positive test was significantly lower but not zero in controls, implying that some patients are colonised without becoming ill. Remarkably, in our cohort, patients with VP had a different clinical presentation and a lower inflammatory response as measured by biomarkers.
These observations support the role of respiratory viruses in CAP, with distinctive clinical and analytical features.
In the present study, sputum samples were used instead of nasopharyngeal sampling. It is more difficult to obtain sputum of good quality than nasopharyngeal swabs, but with sputum, we can overcome, to some extent, the potential colonisation role in the upper respiratory tract and optimise the viral load. Lower respiratory tract specimens have obvious advantages for establishing the cause of pneumonia. However, obtaining reliable specimens that are not contaminated by flora from the upper airway is difficult [22, 23]. The implementation of quantitative tests can shed further light on the relation between colonisation and infection and between virus load and severity [24].
In our cohort, influenza and RSV were the most common cause of VP in adults, as previously reported [25]. RSV is increasingly recognised as a cause of respiratory illness in adults and pulmonary infections are associated with increased mortality [26]. However, rhinovirus was the viral pathogen most frequently identified in co-infections. Rhinovirus has been reported in several outbreaks of severe respiratory disease with fatal pneumonia in elderly residents of nursing homes [27].
Surprisingly, H1N1 was infrequently identified. This could be explained because the peak of the pandemic was already over when the study was initiated. The study began in the 47th week of 2009, which represented the end of the flu pandemic in our area. As expected, a clear seasonal pattern was identified in viral infections [28].
Although we have shown the significant contribution of respiratory viruses in patients presenting with pneumonia, S. pneumoniae remains the leading causative agent.
One of our more relevant findings was that clinical presentation, severity, CRP and PCT values were different between VP and BP. These observations have important clinical implications, as they could help to guide empirical therapy in the absence of modern molecular diagnostics. PCT levels can be useful as a guide to shorter courses of antimicrobial therapy or even to avoid the use of antimicrobial therapy in low-risk patients [29, 30]. Nevertheless, the exact role of PCT in the management of pneumonia is still the subject of ongoing discussion and debate [31].
No clear consensus has been reached about whether patients diagnosed as having viral CAP need to be treated with antibiotics [25]. Some experts recommend that all patients with pneumonia should receive antibiotic treatment, because exclusion of the presence of bacterial co-infection is very difficult, if not impossible. More studies are needed in order to validate these results.
Our study has both strengths and limitations. Very few data are available trying to correlate viral aetiology with clinical, radiological and laboratory features in CAP. The strengths of our study are its prospective nature, the thorough collection of clinical and laboratory data from a cohort of consecutive patients who had been admitted to a single institution and the systematic use of modern molecular methods of diagnosis.
There are also several limitations of the study. First and foremost, despite our best efforts and a detailed study protocol, a number of convalescent serum specimens were missed, thereby, potentially underestimating the number of cases of BP and potentially underestimating the number of co-infections. Second, we excluded patients without sputum because not obtaining specimens for conducting PCR tests was a study protocol violation. Third, we used a qualitative instead of quantitative PCR test. Finally, strictly speaking, a study on ‘confirmed’ VP would require lung tissue samples from all of the enrolled patients. However, while it may be a selection bias in the groups, VP and BP had distinctive clinical and laboratory features concordant with the aetiologic diagnosis, as to allow us to suspect a bacterial versus a viral cause.
To sum up, multiplex PCR testing in CAP patients increases the number of microbiological diagnoses and allows the identification of a viral infection in more than one-third of cases. However, S. pneumoniae still remains the leading cause of CAP. Clinical presentation, severity and CRP and PCT values are different between VP and BP, and should help the clinician to guide empirical therapy.
Acknowledgements
Anna Sangil has no conflict of interest. Esther Calbo has no conflict of interest. Alejandro Robles has no conflict of interest. Susana Benet has no conflict of interest. Maria Eugenia Viladot has no conflict of interest. Vanesa Pascual has no conflict of interest. Eva Cuchi has no conflict of interest. Josefa Perez has no conflict of interest. Bienvenido Barreiro has no conflict of interest. Baltasar Sanchez has no conflict of interest. Juan Torres has no conflict of interest. Lidia Canales has no conflict of interest. Jose Angel De Marcos has no conflict of interest. Javier Garau has been a consultant of GSK, Pfizer, AstraZeneca, Novartis and Bayer AG, and has lectured for Novartis, AstraZeneca, Pfizer and Bayer AG.
Funding
No funding of any kind has been received.
References
- 1.File TM. Community-acquired pneumonia. Lancet. 2003;362:1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garau J, Calbo E. Community-acquired pneumonia. Lancet. 2008;371:455–458. doi: 10.1016/S0140-6736(08)60216-0. [DOI] [PubMed] [Google Scholar]
- 3.Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Templeton KE, Scheltinga SA, Beersma MF, et al. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 6.Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 7.Lim WS, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Templeton KE, Scheltinga SA, van den Eeden WC, et al. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnstone J, Majumdar SR, Fox JD, et al. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest. 2008;134:1141–1148. doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jennings LC, Anderson TP, Beynon KA, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 11.Angeles Marcos M, Camps M, Pumarola T, et al. The role of viruses in the aetiology of community-acquired pneumonia in adults. Antivir Ther. 2006;11:351–359. [PubMed] [Google Scholar]
- 12.Johansson N, Kalin M, Tiveljung-Lindell A, et al. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito A, Kohno S, Matsushima T, et al. Prospective multicenter study of the causative organisms of community-acquired pneumonia in adults in Japan. J Infect Chemother. 2006;12:63–69. doi: 10.1007/s10156-005-0425-8. [DOI] [PubMed] [Google Scholar]
- 14.Charles PG, Whitby M, Fuller AJ, et al. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46:1513–1521. doi: 10.1086/586749. [DOI] [PubMed] [Google Scholar]
- 15.Hohenthal U, Vainionpää R, Meurman O, et al. Aetiological diagnosis of community acquired pneumonia: utility of rapid microbiological methods with respect to disease severity. Scand J Infect Dis. 2008;40:131–138. doi: 10.1080/00365540701534525. [DOI] [PubMed] [Google Scholar]
- 16.Hohenthal U, Vainionpää R, Nikoskelainen J, et al. The role of rhinoviruses and enteroviruses in community acquired pneumonia in adults. Thorax. 2008;63:658–659. doi: 10.1136/thx.2007.093922. [DOI] [PubMed] [Google Scholar]
- 17.Diederen BMW, Van Der Eerden MM, Vlaspolder F, et al. Detection of respiratory viruses and Legionella spp. by real-time polymerase chain reaction in patients with community acquired pneumonia. Scand J Infect Dis. 2009;41:45–50. doi: 10.1080/00365540802448799. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman D, Shimoni A, Shemer-Avni Y, et al. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138:811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalmers JD, Taylor JK, Singanayagam A, et al. Epidemiology, antibiotic therapy, and clinical outcomes in health care-associated pneumonia: a UK cohort study. Clin Infect Dis. 2011;53:107–113. doi: 10.1093/cid/cir274. [DOI] [PubMed] [Google Scholar]
- 20.Johansson N, Kalin M, Hedlund J. Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand J Infect Dis. 2011;43:609–615. doi: 10.3109/00365548.2011.570785. [DOI] [PubMed] [Google Scholar]
- 21.von Baum H, Schweiger B, Welte T, et al. How deadly is seasonal influenza-associated pneumonia? The German Competence Network for Community-Acquired Pneumonia. Eur Respir J. 2011;37:1151–1157. doi: 10.1183/09031936.00037410. [DOI] [PubMed] [Google Scholar]
- 22.Murdoch DR, Anderson TP, Beynon KA, et al. Evaluation of a PCR assay for detection of Streptococcus pneumoniae in respiratory and nonrespiratory samples from adults with community-acquired pneumonia. J Clin Microbiol. 2003;41:63–66. doi: 10.1128/JCM.41.1.63-66.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.File TM, Jr, Marrie TJ. Burden of community-acquired pneumonia in North American adults. Postgrad Med. 2010;122:130–141. doi: 10.3810/pgm.2010.03.2130. [DOI] [PubMed] [Google Scholar]
- 24.Percivalle E, Rovida F, Piralla A, et al. Rapid typing, subtyping and RNA quantification of influenza virus type A strains in respiratory secretions. New Microbiol. 2008;31:319–327. [PubMed] [Google Scholar]
- 25.Marcos MA, Esperatti M, Torres A. Viral pneumonia. Curr Opin Infect Dis. 2009;22:143–147. doi: 10.1097/QCO.0b013e328328cf65. [DOI] [PubMed] [Google Scholar]
- 26.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 27.Longtin J, Marchand-Austin A, Winter AL, et al. Rhinovirus outbreaks in long-term care facilities, Ontario, Canada. Emerg Infect Dis. 2010;16:1463–1465. doi: 10.3201/eid1609.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruuskanen O, Lahti E, Jennings LC, et al. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert DN. Procalcitonin as a biomarker in respiratory tract infection. Clin Infect Dis. 2011;52(Suppl 4):S346–S350. doi: 10.1093/cid/cir050. [DOI] [PubMed] [Google Scholar]
- 30.Schuetz P, Albrich W, Christ-Crain M, et al. Procalcitonin for guidance of antibiotic therapy. Expert Rev Anti Infect Ther. 2010;8:575–587. doi: 10.1586/eri.10.25. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert DN. Use of plasma procalcitonin levels as an adjunct to clinical microbiology. J Clin Microbiol. 2010;48:2325–2329. doi: 10.1128/JCM.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


