Abstract
Human Parainfluenzaviruses (PIVs) account for a significant proportion of viral acute respiratory infections (ARIs) in children, and are also associated with morbidity and mortality in adults, including nosocomial infections. This work aims to describe PIV genotypes and their clinical and epidemiological distribution. Between December 2016 and December 2017, 6121 samples were collected, and submitted to viral culture and genomic quantification, specifically Parainfluenza 1–4 (PIV1–4), Influenza A and B, Respiratory Syncytial Virus (RSV) A and B, Adenovirus, Metapneumovirus, Coronavirus, Rhinovirus, and Enterovirus. Normalized viral load, as (log10) copies/103 cells, was calculated as virus Ct, determined by multiple qRT-PCR, as a function of the Ct of β-globin. PIV was confirmed in 268 cases (4.37%), and linked to both upper and lower respiratory tract disease, being more frequent in children than in adults (5.23 and 2.43%, respectively). PIV1 and PIV3 were most common (31 and 32.5%, of total PIV positive samples, respectively), with distribution being similar in children and adults, as was viral load. PIV type was correlated with seasonality: PIV3 being more frequent in winter and spring, PIV1 in summer, and PIV 4 in fall. No correlation between vial load and clinical severity was found. Novel findings were that PIV viral load was higher in fall than in other seasons, and PIV4, classically linked to mild respiratory symptoms, was circulating, in children and adults, at all levels of symptoms throughout the year.
Keywords: Parainfluenzavirus, Season, Acute respiratory infections, Viral load
Background
Viruses are the predominant cause of acute respiratory infection worldwide, being responsible for a high rate of morbidity and mortality especially in children, the elderly, and immunosuppressed patients [1–3].
Human Parainfluenzaviruses (PIVs) account for a significant proportion of viral acute respiratory infections (ARIs) in children, representing the second most common cause of upper and lower respiratory tract infections, behind Respiratory Syncytial Virus (RSV) [4]. Both PIVs and RSV have also been associated with morbidity and mortality in adults, including nosocomial infections [5].
There are four known types of PIV; PIV types 1 and 3 belong to the genus Respirovirus, while types 2 and 4 are from the genus Rubulavirus [6–8].
However, little information on the epidemiology and clinical characteristics of the different types of PIV infection has been published, especially works that consider adults and children separately.
A prospective, descriptive study was carried out in order to better characterize the different PIV types and their clinical features in children and adults with acute respiratory infection.
Study design
Patients and samples
Between December 2016 and December 2017, 6121 respiratory samples (3689 pharyngeal, 137 nasal swabs, 2027 nasopharyngeal swabs, 160 low respiratory tract samples, and 108 from nasopharyngeal washing) were collected and taken, in viral transport medium (ViralPack, Biomedics, Spain) to the Clinical Virology Laboratory of the Hospital Universitario Central de Asturias (HUCA). Samples were from 4264 children (< 16 years) and 1845 adults (aged > 65), of whom 328 were immunosuppressed (oncology or hematology patients). Symptomatology of the samples corresponded to 1926 upper respiratory tract infections (URTIs), 681 lower respiratory tract infections (LRTIs), and 317 asthma cases and 3197 systemic symptoms (febrile syndrome, lymphadenopathies, and other respiratory infections not classified as URTI/LRTI).
Laboratory diagnosis
The samples were processed following laboratory protocols and were each then divided into two aliquots. The first (1 ml) was used for conventional monolayer cell culture (MRC-5 and mix of LLC-MK2, A549 and Hep-2), while the second (500 μl) was used for nucleic acid extraction. Quantitative real-time reverse transcription PCR (qRT-PCR) analyses were performed using type-specific primer pairs and MGB probes designed to target either the conserved regions of the matrix gene of PIV1/3 or of the polymerase gene of PIV2/4 in order to identify the strains of PIV present in the various respiratory samples.
Genomic detection, typing, and viral load
Nucleic acids were extracted using the automated purifier MagNAPure 96 (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s instructions. The extracted nucleic acids were then resuspended in a final volume of 70 μl.
PIV1–4 genome was detected by a multiplex qRT-PCR for Parainfluenzavirus genus, along with Metapneumovirus and Coronavirus genus (as per the protocol of the laboratory). Briefly, 5 μl of extracted nucleic acid was amplified, in a final volume of 10 μl, with TaqMan Fast 1-Step Master Mix (Life technologies, CA), four PIV primers (Thermo Fisher Scientific, MA), and two PIV FAM-labeled MGB probes (ABI, CA) (Table 1). qRT-PCR amplification comprised an initial retrotranscription at 45 °C for 15 min, followed by a denaturation cycle at 95 °C for 10 min, and 40 amplification cycles consisting of 95 °C for 10 s and annealing/extension at 60 °C for 30 s. Amplifications were achieved in a 7300 or 7500 Real Time PCR System (Applied Biosystems).
Table 1.
Parainfluenzavirus (PIV) primer and probe sequences
| Virus | Gene | Primer | Sequence (5′-3′) | Size | MGB probe | Sequence (5′-3′) |
|---|---|---|---|---|---|---|
| PIV1 | Matrix | Para1-S | CATGTACATGCAACAGGCACATTA | 123 pb | Para13-FAM | TTTGTTATCCYTTAATGGAT |
| Para1-A | TGAAGCTGCCCAGATGACTAGA | |||||
| PIV3 | Para3-S | CATGTCAATGCAACTGGATCCATA | 123 pb | |||
| Para3-A | TGATGAAGCCCAGATAACCAGA | |||||
| PIV2 | Polymerase | Para2/4-S | TATGTWGGTGAYCCRTTYAATCCT | 164 pb | Para24-FAM | GGAGGTATYGAAGGWCT |
| Para2-A | GCTGAAGAGAGGATGATCACAGA | |||||
| PIV4 | Para2/4-S | TATGTWGGTGAYCCRTTYAATCCT | 175 pb | |||
| Para4-A | TCTTAGATTCTGCAGATGACAAYAA |
A standard curve was performed using serial dilutions from 10 to 1010 of amplicon-based positive controls of each type of PIV to determine the sensitivity of the assay. The limit of detection was 10 copies. In any run, positive controls were included and the Ct had to be between 27 and 33: 30–33 for PIV-1, 28–31 for PIV2, 27–30 for PIV3, and 28–31 for PIV4.
The specificity was evaluated using viral strains frequently found in human respiratory infections such as Influenza types A and B, Adenovirus, Metapneumovirus, Coronavirus, Rhinovirus, and Enterovirus as well as other viruses like CMV, EBV, HSV1, VZV, HHV6, HHV7, or Parvovirus.
Viral and cell quantification were performed by interpolating Ct data from qPCR assays of β-globin and of each virus genotype in a standard curve, as described previously [9, 10]. Viral load was calculated as the ratio of copies of viral genome/number of cells and expressed as (log10) copies/103 cells. Infection and viral load data was then compared by age group, clinical manifestations, and seasonality.
Four different qRT-PCR assays, which included the FAM-labeled MGB probe and separate primers for PIV1, PIV2, PIV3, and PIV4 (Table 1), were carried out to characterize Parainfluenzavirus type.
Statistical analyses
Statistical tests were performed using GraphPad InStat v.3. for Windows 2007 (GraphPad Software, USA). Tests were considered significant when p value was < 0.05.
Results
Demographic data
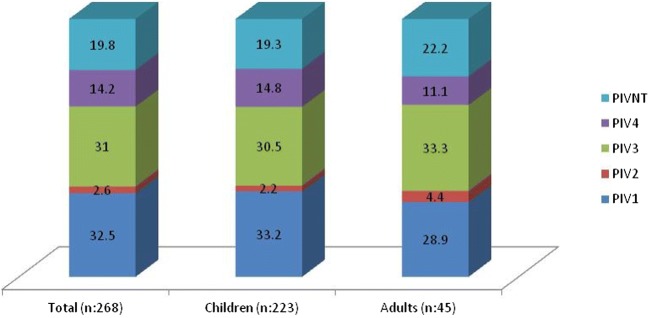
Of the samples from 6121 patients included in the study, viruses were detected in 2326. Enterovirus was the most frequently detected viral agent 742 (12.1%), followed by Adenovirus 621 (10.14%). Less frequent were Rhinovirus in 285 samples (4.6%), RSV in 242 (3.9%), Coronavirus in 178 (2.9%), and Influenza A (IA) in 143 (2.3%). Influenza B (B) was detected in 75 (1.2%) and Metapneumovirus in 40 (0.65%). Parainfluenzavirus infection was confirmed in 268 (4.37%) samples, a similar rate to RSV, of which 223 (5.23%) corresponded to samples from children and 45 (2.43%) from adults (p < 0.0001) (26 (2.46%) aged between 16 and 65 and 19 (2.5%) older than 65) (Table 2 and Figs. 1 and 2). Furthermore, 17 (5.18%) of the 328 immunosuppressed patients tested positive, a rate similar to that in the child population (p = 0.44).
Table 2.
Incidence and distribution of Parainfluenzavirus by type, age, and viral load
| Total (n:268) | VL | Children (n:223) | Adults (n:45) | p | |
|---|---|---|---|---|---|
| Incidence | 4.37% | 5.23% | 2.43% | < 0.0001 | |
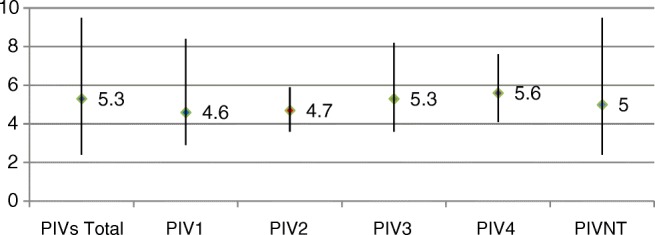
| VL | 5.3 ± 1.2 (2.4–9.5) | 5.3 ± 1.5 (2.4–9.5) | 5.6 ± 1.3 (2.6–8.4) | ns | |
| PIV1 | 87 (32.5%) | 4.6 ± 1.3 (2.9–8.4) | 74 (33.2%) | 13 (28.9%) | ns |
| PIV2 | 7 (2.6%) | 4.7 ± 0.9 (3.6–5.9) | 5 (2.2%) | 2 (4.4%) | ns |
| PIV3 | 83 (31%) | 5.3 ± 1 (3.6–8.2) | 68 (30.5%) | 15 (33.3%) | ns |
| PIV4 | 38 (14.2%) | 5.6 ± 0.8 (4.1–7.6) | 33 (14.8%) | 5 (11.1%) | ns |
| PIVNT | 53 (19.8%) | 5 ± 1.4 (2.4–9.5) | 43 (19.3%) | 10 (22.2%) | ns |
| p | < 0.0001 | ns | ns | ns |
VL viral load (log10) copies/103 cells, NT not typed
Fig. 1.
Distribution (percentage) of PIV type according to age
Fig. 2.
Viral load of the different types of Parainfluenzavirus
PIV1/PIV3 were more frequent than PIV4/PIV2 (found in 32.5/31% compared to 14.2/2.6% of cases, respectively), although the distribution of the various types was similar in children and adults, as were viral loads (p = ns) (Table 3). No patients were found to have more than one type of PIV.
Table 3.
Distribution of types of PIV by season, showing viral load for each season
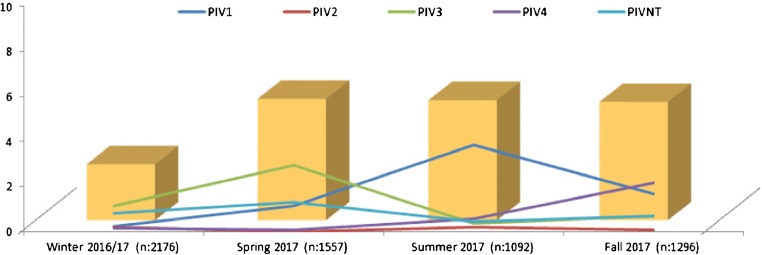
| Winter 2016/2017 (n:2176) | Spring 2017 (n:1557) | Summer 2017 (n:1092) | Fall 2017 (n:1296) | p | |
|---|---|---|---|---|---|
| PIV1 | 5 (0.23%) | 18 (1.15%) | 42 (3.86%) | 22 (1.69%) | < 0.0001 |
| PIV2 | 4 (0.18%) | 0 | 2 (0.18%) | 1 (0.08%) | ns |
| PIV3 | 25 (1.15%) | 46 (2.95%) | 3 (0.27%) | 9 (0.69%) | < 0.004 |
| PIV4 | 3 (0.14%) | 1 (0.06%) | 6 (0.55%) | 28 (2.16%) | < 0.002 |
| PIVNT | 18 (0.82%) | 20 (1.28%) | 6 (0.55%) | 9 (0.69%) | ns |
| Total | 55 (2.52%) | 85 (5.46%) | 59 (5.4%) | 69 (5.32%) | < 0.0001 |
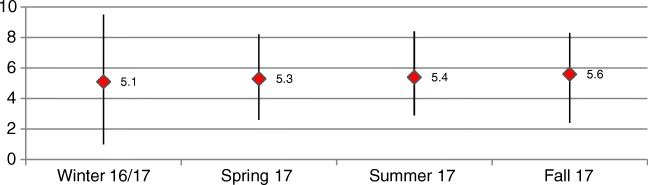
| VL | 5.1 ± 1.2 (2.9–9.5) | 5.3 ± 1 (2.6–8.2) | 5.4 ± 1.3 (2.9–8.4) | 5.6 ± 1.1 (2.4–8.3) | < 0.0001 |
VL viral load (log10) copies/103 cells, NT not typed
The genotype detected in the 17 immunosuppressed patients with PIV infection, were 8 (47.06%) cases of PIV3, 7 (41.17%) of PIV1, 1 (5.88%) of PIV2, and 1 of (5.88%) PIV4.
Seasonality
Distribution and viral load of PIV-positive samples are shown in Table 3 and Figs. 3 and 4.
Fig. 3.
Distribution of PIV types across seasons
Fig. 4.
PIV viral load by season
As Table 3 demonstrates, the season with the lowest levels of circulating Parainfluenzavirus is during winter (p < 0.0001). In terms of genotypes, positive PIV3 samples were far more frequent in winter and spring, while a peak in PIV1-infected patients was noted in summer and PIV4 cases were significantly more common in fall, when, in addition, overall PIV viral load was higher than in other seasons (p < 0.0001).
Clinical diagnosis
Table 4 shows PIV infection rate and viral load by clinical status for each PIV type. No single clinical status was more predominantly associated with PIV infection: Parainfluenzavirus was detected in 103 (5.35%) cases of URTI, 34 (4.99%) cases of LRTI, 12 (3.78%) of asthma, and in 119 (3.72%) cases presenting systemic symptoms (febrile syndrome, lymphadenopathies, and other cases not classified as URTI/LRTI) (p = ns).
Table 4.
Clinical diagnosis and viral load of patients with Parainfluenzavirus infection
| URTI (n:1926) | LRTI (n:681) | Asthma (n:317) | Systemic symptoms (n:3197) | p | |
|---|---|---|---|---|---|
| PIV 1 | 34 (1.77%) | 8 (1.17%) | 4 (1.26%) | 41 (1.28%) | ns |
| PIV 2 | 2 (0.1%) | 1 (0.15%) | 1 (0.32%) | 3 (0.09%) | ns |
| PIV 3 | 39 (2.02%) | 10 (1.47%) | 3 (0.95%) | 31 (0.97%) | ns |
| PIV 4 | 14 (0.73%) | 7 (1.03%) | 1 (0.32%) | 16 (0.5%) | ns |
| PIV NT | 14 (0.73%) | 8 (1.17%) | 3 (0.95%) | 28 (0.87%) | ns |
| Total PIV | 103 (5.35%) | 34 (4.99%) | 12 (3.78%) | 119 (3.72%) | ns |
| VL | 5.35 ± 1.17 (3.1–9.5) | 5.1 ± 1.2 (2.9–7.6) | 5.33 ± 1.3 (2.9–7.9) | 5.38 ± 1.19 (2.4–8.4) | ns |
VL viral load (log10) copies/103 cells, NT not typed
Infection rates ranged from 3.72% for systemic symptoms to 5.35% in cases of URTI (p = ns) and viral load varied between 5.1 (log10) copies/103 cells in LRTI and 5.38 (log10) copies/103 cells in systemic symptoms (p = ns).
Discussion
Respiratory infections are a major health challenge around the world. The causative agent in 80% of such infections are viruses, including PIV [11]. Although PIVs are commonly recognized as a significant cause of morbidity and mortality in children, their impact in adults is less well characterized [12]. In adults, PIVs usually cause mild URTIs, although they can also lead to life-threatening LRTIs and death in transplant recipients, as well as nosocomial infections and nursing-home outbreaks [13], similar to the Influenza virus [14–17].
The efficacy of Ribavirin in the treatment of PIVs is limited and can be contradictory [18]. Therefore, the early and accurate diagnosis of the viral agent continues to be a priority in order to prevent secondary bacterial infection, which increases morbidity and mortality, as well as the unnecessary use of antibiotics [19].
In the present work, the most frequently found viruses were Enterovirus and Adenovirus with rates of 12.1 and 10.14%, respectively. The prevalence of PIVs was 4.37%, similar to Rhinovirus (4.6%) and RSV (3.9%). Children had significantly higher rate of infection than adults, although the lower rate in adults is in line with the rates of infection of the other viruses studied here. The rate of infection among immunosuppressed patients was found to be very similar to that of children.
In terms of PIV type, our results show that PIV1 and PIV3 were the most frequent genotypes. However, PIV4 in our study was more frequent than PIV2, a result not observed in the US study. The same distribution pattern for the different PIV genotypes was found in both adults and children, as well as in immunosuppressed patients, as also found in a study of adults and children in the USA between 1990 and 2004 [13].
PIVs have been estimated to cause 10% of ARIs during the winter [20–22]. However, the results of our study indicate that in fact, for this type of clinical manifestation, PIV is circulating at lower rates (approximately 2%) in winter than during the rest of the year (approximately 5%). In addition, while it has been described in the literature that clinical manifestations of PIV vary depending upon the viral genotype involved, our findings do not support this. We did however find evidence of seasonal differences in the incidence of PIV genotypes: PIV3 being more common in spring, and winter, PIV1 in summer and PIV4 in fall, with PIV2 being present at only very lower rates in all seasons except spring, when it is totally absent. These findings are in contrast to those of other studies where no such seasonal effect has been observed for PIV4 since historically this genotype was considered to only be associated with mild respiratory problems and as such was not included in the diagnosis of ARIs.
Despite associations between PIV4 and ARIs having been proposed for more than half a century, it is not commonly included in clinical diagnoses, although some studies do highlight its potential virulence [23, 24, 25]. We, however, found this virus to be circulating among both pediatric and adult patients presenting with different types of symptoms across the entire period tested, along with other PIV types, with PIV4 occurrence peaking in fall.
However, the most novel, and controversial in terms of diagnostics, finding from our study relates to viral load. The results provide evidence that in fall PIVs are circulating with a higher viral load than at other times in the year. As shown in Table 2, in fall, while it is not significant, the viral load of PIV4 is slightly higher than the other genotypes. At the same time, rate of infection of PIV4 is significantly higher in fall. These two factors, greater incidence of PIV4 and its concomitant higher viral load, may well be the explanation for this interesting novel finding. As well as needing to corroborate these findings, further studies, with a greater number of samples, are also required to examine the possible link between viral load and the severity of clinical symptoms.
In addition, the results of this work provide evidence for the same rates of PIV infection in mild URTIs and LRTIs, as well as evidencing that clinical manifestations are not dependent on viral genotype or a specific viral load, thus distinguishing it from other studies [26, 27], something which could perhaps be corroborated with a greater number of patients.
In summary, this study provides evidence that PIVs cause respiratory infections in all age groups, albeit that, as has been found in other studies, children are more commonly affected, and we also show that virus replication is more efficient in fall. It is, however, important to note that, at least in our study scenario, PIVs are not only related to mild infections, such as those of the upper respiratory tract. In addition, it was observed that PIV4, on which there are very few studies because of the assumption it is related only to mild symptoms, was not only frequent, but was also associated with lower respiratory tract infections. The results presented here support the notion that testing for PIV type could provide additional valuable information as regards the distribution of the genotypes in terms of age and pathology, as well as seasonal differences in these data.
Acknowledgements
We thank Ronnie Lendrum for her help with correcting the English.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethics approval
This study was approved by the Hospital Universitario Central de Asturias (HUCA) Ethical Committee.
Contributor Information
Marta E. Álvarez-Argüelles, Phone: +34 985108720, Email: martaealvarez@gmail.com
Susana Rojo-Alba, Email: ssnrj4@gmail.com.
Zulema Pérez Martínez, Email: zulema.pm@hotmail.com.
Álvaro Leal Negredo, Email: alvaro.lene.21@gmail.com.
José Antonio Boga Riveiro, Email: joseantonio.boga@sespa.es.
María Agustina Alonso Álvarez, Email: mariaagustina.alonso@sespa.es.
Julián Rodríguez Súarez, Email: julian.rodriguez@sespa.es.
María de Oña Navarro, Email: maria.deona85@gmail.com.
Santiago Melón García, Email: santiago.melon@sespa.es.
References
- 1.Clark TW, Medina MJ, Batham S, Parmar S, Nicholson KG. Adults hospitalised with acute respiratory illness rarely have detectable bacteria in the absence of COPD or pneumonia; viral infection predominates in a large prospective UK sample. J Inf Secur. 2014;69:507–515. doi: 10.1016/j.jinf.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, Chappell JD, Qi C, Hart EM, Carroll F, Trabue C, Donnelly HK, Williams DJ, Zhu Y, Arnold SR, Ampofo K, Waterer GW, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, McCullers JA, Pavia AT, Edwards KM, Finelli L, Study Team CDCEPIC. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman S. Epidemiology of acute respiratory infections in children of developing countries. Rev Infect Dis. 1991;13(Suppl 6):S454–S462. doi: 10.1093/clinids/13.Supplement_6.S454. [DOI] [PubMed] [Google Scholar]
- 4.Pecchini R, Berezin EN, Souza MC, Vaz-de-Lima Lde A, Sato N, Salgado M, Ueda M, Passos SD, Rangel R, Catebelota A. Parainfluenza virus as a cause of acute respiratory infection in hospitalized children. Braz J Infect Dis. 2015;19(4):358–362. doi: 10.1016/j.bjid.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spahr Y, Tschudin-Sutter S, Baettig V, Compagno F, Tamm M, Halter J, Gerull S, Passweg J, Hirsch HH, Khanna N. Community-acquired respiratory Paramyxovirus infection after allogeneic hematopoietic cell transplantation: a single-center experience. Open Forum Infect Dis. 2018;5(5):ofy077. doi: 10.1093/ofid/ofy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villaran MV, Garcia J, Gomez J, Arango AE, Gonzales M, Chicaiza W, Alemán W, Lorenzana de Rivera I, Sanchez F, Aguayo N, Kochel TJ, Halsey ES. Human parainfluenza virus in patients with influenza-like illness from central and South America during 2006–2010. Influenza Other Respir Viruses. 2014;8(2):217–227. doi: 10.1111/irv.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilar JC, Perez-Brena MP, Garcia ML, Cruz N, Erdman DD, Echevarria JE. Detection and identifi cation of human parainfl uenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription-PCR. J Clin Microbiol. 2000;38(3):1191–1195. doi: 10.1128/jcm.38.3.1191-1195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao N, Ji Y, Xie Z, Wang H, Wang H, An J, Zhang X, Zhang Y, Zhu Z, Cui A, Xu S, Shen K, Liu C, Yang W, Xu W. Human parainfl uenza virus-associated respiratory tract infection among children and genetic analysis of HPIV-3 strains in Beijing, China. PLoS One. 2012;7(8):e43893. doi: 10.1371/journal.pone.0043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Álvarez-Argüelles ME, de Oña-Navarro M, Rojo-Alba S, Torrens-Muns M, Junquera-Llaneza ML, Antonio-Boga J, Pérez-Castro S, Melón-García S. Quantification of human papilloma virus (HPV) DNA using the Cobas 4800 system in women with and without pathological alterations attributable to the virus. J Virol Methods. 2015;222:95–102. doi: 10.1016/j.jviromet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Novo M, Boga JA, Álvarez-Argüelles ME, Rojo-Alba S, Fernández A, Menéndez MJ, de Oña M, Melón S. Human respiratory syncytial virus load normalized by cell quantification as predictor of acute respiratory tract infection. J Med Virol. 2018;90(5):861–866. doi: 10.1002/jmv.25020. [DOI] [PubMed] [Google Scholar]
- 11.Abedi GR, Prill MM, Langley GE, Wikswo ME, Weinberg GA, Curns AT, Schneider E. Estimates of Parainfl uenza virus-associated hospitalizations and cost among children aged less than 5 years in the United States, 1998–2010. J Pediatric Infect Dis Soc. 2016;5(1):7–13. doi: 10.1093/jpids/piu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001;344:1917. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 13.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. Seasonal trends of human parainfluenza viral infections: United States, 1990-2004. Clin Infect Dis. 2006;43:1016. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 14.Ison MG. Respiratory viral infections in transplant recipients. Antivir Ther. 2007;12:627. [PubMed] [Google Scholar]
- 15.Russell E, Ison MG. Parainfluenza virus in the hospitalized adult. Clin Infect Dis. 2017;65:1570. doi: 10.1093/cid/cix528. [DOI] [PubMed] [Google Scholar]
- 16.Whimbey E, Champlin RE, Couch RB, Englund JA, Goodrich JM, Raad I, Przepiorka D, Lewis VA, Mirza N. Other authors. Community respiratory virus infections among hospitalized adult bone marrow transplant recipients. Clin Infect Dis. 1996;22:778–782. doi: 10.1093/clinids/22.5.778. [DOI] [PubMed] [Google Scholar]
- 17.Glasgow KW, Tamblyn SE, Blair G. A respiratory outbreak due to parainfluenza virus type 3 in a home for the aged – Ontario. Can Commun Dis Rep. 1995;21:57–61. [PubMed] [Google Scholar]
- 18.Park SY, Baek S, Lee SO, Choi SH, Kim YS, Woo JH, Sung H, Kim MN, Kim DY, Lee JH, Lee JH, Lee KH, Kim SH. Efficacy of oral ribavirin in hematologic disease patients with paramyxovirus infection: analytic strategy using propensity scores. Antimicrob Agents Chemother. 2013;57(2):983–989. doi: 10.1128/AAC.01961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhary ML, Anand SP, Heydari M, Rane G, Potdar VA, Chadha MS, Mishra AC. Development of a multiplex one step RT-PCR that detects eighteen respiratory viruses in clinical specimens and comparison with real time RT-PCR. J Virol Methods. 2013;189(1):15–19. doi: 10.1016/j.jviromet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Côté S, Peret TC, Erdman DD, Anderson LJ. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis. 2002;186:1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- 21.Henderson FW. Pulmonary infections with respiratory syncytial virus and the parainfluenza viruses. Semin Respir Infect. 1997;2:112–121. [PubMed] [Google Scholar]
- 22.Sharova NK, Kubinova I, Vorkunova GK, Tumova B, Bukrinskaia AG. Conservativism of the matrix protein of paramyxoviruses. Vopr Virusol. 1989;34:573–575. [PubMed] [Google Scholar]
- 23.Rodríguez-Serrano DA, Nieto-Cabrera M, Conesa J, Culebras-López E. Neumonía por virus parainfluenza tipo 4 y púrpura trombótica trombocitopénica. Med Int. 2012;36(3):235. doi: 10.1016/j.medin.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFarlane HJ, MacDonald J, Collins TC, Molyneaux PJ, Carman WF. Severe pneumonia after cardiac surgery as a result of infection with parainfluenza virus type 4. J Cardiothorac Vasc Anesth. 2009;23:84–86. doi: 10.1053/j.jvca.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Lau SK, Li KS, Chau KY, So LY, Lee RA, Lau LY. Clinical and molecular epidemiology of human parainfluenza virus 4 infections in Hong Kong: subtype 4B as common as subtype 4A. J Clin Microbiol. 2009;47:1549–1552. doi: 10.1128/JCM.00047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright PF. Parainfluenza viruses. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. 6. Philadelphia: Elsevier Churchill Livingstone; 2005. p. 1998. [Google Scholar]
- 27.Kim YJ, Boeckh M, Englund JA. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin Respir Crit Care Med. 2007;28:222. doi: 10.1055/s-2007-976494. [DOI] [PubMed] [Google Scholar]