Abstract
Myxovirus resistance A (MxA) is an antiviral protein induced by type I interferons α and β (IFN-α and IFN-β) that can inhibit virus replication. We examined whether the MxA polymorphisms were related to the risk and severity of enterovirus 71 (EV71) infection in Chinese populations. The MxA C-123A and G-88T polymorphisms were genotyped in two independent case–control populations in China by polymerase chain reaction-based restriction fragment length polymorphism (PCR-RFLP) analysis. Multivariate logistic regression analysis was used to calculate adjusted odds ratios (ORs) and 95 % confidence intervals (95 % CIs). MxA messenger RNA was quantified by real-time quantitative PCR in peripheral blood mononuclear cells (PBMCs) from 45 healthy children and 19 patients with EV71 infection. Significantly decreased susceptibility to EV71 infection was observed for the -123A allele and -88T allele carriers, with ORs (95 % CIs) estimated as 0.56 (0.39–0.81) and 0.64 (0.47–0.88), respectively, in the northern population. This association was confirmed in the southern population, with ORs (95 % CIs) estimated as 0.58 (0.38–0.89) and 0.67(0.47–0.95), respectively. The A- 123T- 88 haplotype was also significantly associated with lower risk of EV71 infection in both the northern (OR = 0.62; 95 % CI = 0.44–0.85) and the southern population (OR = 0.63; 95 % CI = 0.43–0.92). Furthermore, we observed higher MxA messenger RNA levels in IFNβ1a-stimulated PBMCs from the -123A or -88T allele carriers compared with that from nocarriers. Our findings suggest that polymorphisms in the MxA promoter may play a role in mediating the susceptibility to EV71 infection in Chinese population.
Keywords: Allele Carrier, Southern Population, EV71 Infection, Northern Population, EV71 Outbreak
Introduction
Enterovirus 71 (EV71) is positive-stranded RNA virus belonging to the enterovirus genus of the Picornauiridae family. EV71 has been regarded as the most important neurotropic enterovirus after the primary eradication of the poliovirus. There is no specific antiviral therapy or vaccine for EV71. Since its discovery in 1969, EV71 outbreaks have been reported periodically throughout the world. The size and frequency of EV71 outbreaks have greatly increased over the past decade in the Asia–Pacific region, especially in southeast Asia (Solomon et al. 2010). In mainland China, EV71 infection has become more and more severe and led to 509 and 569 deaths in 2011 and 2012, respectively, according to the report of Ministry of Health, China.
Individual susceptibility to EV71 infection seems to be variable (Chang et al. 2008; Yang et al. 2001, 2012). Patients with EV71 infection displayed wide clinical spectrum, ranging from asymptomatic or mild hand-foot-mouth disease (HFMD) to severe neurological disease. Similar to other infectious diseases, EV71 infection may also be the result of complex interactions between pathogen, environmental factors and host genes. The identification of susceptibility genes contributing to EV71 infection would assist in predicting individual and population risks of EV71 infection, as well as clarifying pathogenesis of this infectious disease.
Type I interferons α and β (IFN-α and IFN-β) system is a key component of the innate immune response and the first line of defense against viral infection including EV71 infection (Liu et al. 2005; Yi et al. 2011). The Myxovirus resistance (Mx) proteins are dynamin-like GTPases that are induced by IFN-α and IFN-β and have antiviral activity against RNA viruses (Sadler and Williams 2008). In humans, two distinct Mx (MxA and MxB) are encoded on chromosome 21, while only MxA has exhibited antiviral activity (Sadler and Williams 2008). Transfected cells (Chieux et al. 2001; Kochs and Haller 1999) and transgenic mice (Pavlovic et al. 1995) overexpressing MxA acquire a high degree of antiviral resistance against several RNA viruses. In humans, synthesis of MxA protein is induced during viral infections and may protect humans from infection (Roers et al. 1994; von Wussow et al. 1990). As for EV71, it has been shown that type I IFN subtypes can inhibit EV71 replication and the antiviral effect was caused by a strong induction of the downstream antiviral effectors especially MxA (Yi et al. 2011). On the basis of the functional role of MxA in the pathogenesis of viral infections in vivo and in vitro, we hypothesize that MxA might be a potential candidate susceptibility gene for EV71 infection.
Among the validated polymorphisms in MxA gene, two functional single nucleotide polymorphisms (SNPs) (C-123A, rs17000900; G-88T, rs2071430) in promoter region have been extensively studied. The electrophoretic mobility shift assays shown that these two polymorphisms could influence the binding of nuclear protein(s) (Ching et al. 2010). Furthermore, by luciferase reporter assay, the basal promoter activity was demonstrated to be higher in promoter constructs containing the minor alleles of the two polymorphisms (Furuyama et al. 2006; Hijikata et al. 2001; Torisu et al. 2004). We and other investigators have reported the association between the aforementioned MxA functional SNPs and risk of infections of viruses, including severe acute respiratory syndrome-coronavirus (SARS-CoV) (Ching et al. 2010; Hamano et al. 2005; He et al. 2006), hepatitis B virus (HBV) (Cao et al. 2009), hepatitis C virus (HCV) (Hijikata et al. 2000) and measles virus (Torisu et al. 2004). The role of the MxA polymorphisms in EV71 infection, however, has never been investigated. In the present study, we examined whether polymorphisms in the MxA gene were related to the risk and severity of EV71 infection among Chinese populations.
Patients and methods
Study population
Patients were recruited between April and November 2010 from 7 hospitals located in either northern or southern China. All the hospitals were surveillance sentinels which were set by China CDC. HFMD was clinically defined as a patient having oral ulcers and vesicular rash on the hands, feet, knees, or buttocks. Complicated patients were defined as cases with central nervous system (CNS) involvement (including encephalitis, poliomyelitis-like syndrome, and encephalomyelitis), myocarditis or pulmonary edema/hemorrhage. In each hospital, all patients who met the clinical definition and willing to participate into the study were recruited.
Stool samples were collected for EV71 RNA detection by real-time PCR using previously described primers (Verstrepen et al. 2002). Viral isolation was performed as described previously (Chen et al. 2010). Serum samples were collected to test the IgM antibody against EV71 by a commercial enzyme-linked immunosorbent assay (ELISA) kit (Beijing Wantai Biological Pharmacy Enterprise Co, Ltd, China). Confirmation of EV71 infections was recognized by one of the following criteria: isolation of EV71 from stool specimens; positive EV71 viral RNA quantitation or positive EV71 IgM results during acute phase of infection.
The controls were randomly selected from healthy children who underwent routine physical examination in the same region during the same period that the cases were recruited. After interviewing their guardians, healthy children with no self-reported or medical-recorded history of HFMD or herpangina were recruited. Their sera samples were collected for EV71-specific IgG antibody test by the Diagnostic Kit for IgG Antibody to Human EV71 (Beijing Beier Bioengineering Co, Ltd, China). Only those with negative EV71 IgG results were recruited as eligible controls. By checking the medical records or by interviewing the participants’ guardians, we determined that all cases and controls were genetically unrelated Han Chinese.
For MxA expression assay, blood samples were obtained from a group of 20 healthy children (13 males and 7 females) aged 26–50 months (mean ± SD, 39.6 ± 6.6) from the northern population, 25 healthy children (16 males and 9 females) aged 24–52 months (mean ± SD, 38.2 ± 8.4) from southern population, and 19 patients with EV71 infection (12 males and 7 females) aged 24–50 months (mean ± SD, 37.8 ± 7.8). At recruitment, informed consent was obtained from all participants’ guardians, and personal information on demographic factors and medical history were collected via structured questionnaire. This study was performed with the approval of the Ethical Committee of Beijing Institute of Microbiology and Epidemiology and conducted according to the principles expressed in the Declaration of Helsinki.
Polymorphism genotyping
Genomic DNA was extracted from peripheral blood by Gene Relax DNA extraction kit (TianGen, Beijing, China) according to the manufacturer’s instructions. The polymorphisms were genotyped using polymerase chain reaction-based restriction fragment length polymorphism (PCR-RFLP) analysis. The primers 5′-GGGAAGGACATGCCTAGGTTC-3′ and 5′-CCGCGAAGAAATGAAACTCAC-3′ were used for amplifying the target region. The PCR conditions were performed as described previously except for an annealing temperature of 57.5 °C (Zhang et al. 2012). The reaction yielded a 263-base pair amplicon. PCR products of 3 μL were then digested with 4U of PstI and 4U HhaI (Takara BioTech, Dalian, China) for the C-123A and G-88T polymorphisms, respectively. Genotyping was done by staff blinded to the subjects’ case/control status. The accuracy of genotyping data obtained from PCR-RFLP analyses was validated by direct sequencing of a 15 % masked, randomly selected sample of cases and controls, and the results were 100 % concordant (Fig. 1).
Fig. 1.
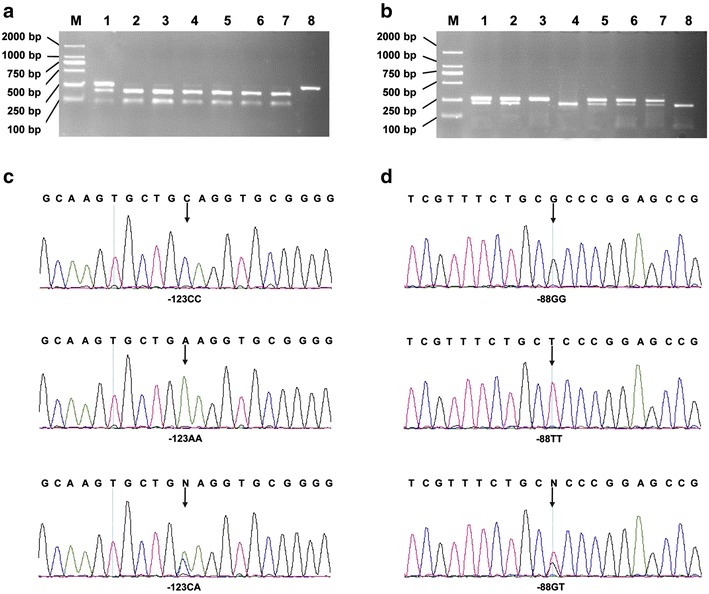
Analysis of the MxA C-123A and G-88T polymorphisms. a PCR-based RFLP analysis for the C-123A polymorphism. Lanes 2–7 were CC genotypes; lane 1 was CA genotype; lane 8 was AA genotype; M was DL2000 marker. b PCR-based RFLP analysis for the G-88T polymorphism. Lanes 4 and 8 were GG genotypes; lanes 1, 2, and 5–7 were GT genotypes; lane 3 was TT genotype; M was DL2000 marker. c Sequencing analysis for genotypes of the C-123A polymorphism. The 3 charts represent the CC, CA and AA genotypes, respectively. d Sequencing analysis for genotypes of the G-88T polymorphism. The 3 charts represent the GG, GT and TT genotypes, respectively. The arrows localize the base changes at the nucleotide positions
Real-time analysis of MxA mRNA
Total RNA of peripheral blood mononuclear cells (PBMCs) from patients with EV71 infection and IFNβ1a (1,000 U/mL, R&D Systems) stimulated PBMCs from healthy children were extracted using Trizol reagent (Invitrogen, USA). Reverse transcription was done using reverse transcriptase system (Promega, USA) according to the manufacturer’s instructions. Quantitative real-time PCR for MxA, with GAPDH as an internal reference gene, was performed using the Roche Light Cycler 2.0 (Roche Applied Science, USA) based on the SYBR-Green method. The primers used for MxA (Fernandez-Arcas et al. 2004) were 5′-ACAGCAAATCAAGGCACTGGA-3′ and 5′-CGGATCAGCTTCTCACCTTCTC-3′and for GAPDH (Xie et al. 2011) were 5′-GGGAAGGTGAAGGTCGGAGT-3′ and 5′-TTGAGGTCAATGAAGGGGTCA-3′. The PCR reaction mixture consisted of 0.1 μmol/L of each primer, 1 X SYBR Green Master mix (Takara), and 50 ng complementary DNA to a final volume of 20 μL. Cycling conditions were 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s and 57 °C for 15 s. Data were analyzed using the 2−∆∆Ct method (Livak and Schmittgen 2001). All analyses were performed in a blinded fashion with the laboratory persons unaware of genotyping data.
Statistical analysis
Genotype and allele frequencies for each polymorphism were determined by gene counting. The fitness to Hardy–Weinberg equilibrium was tested using the random-permutation procedure implemented in the Arlequin package (http://lgb.unige.ch/arlequin). Associations between polymorphisms and risk of EV71 infection were estimated by use of logistic regression analyses. Odds ratios (ORs) and 95 % confidence intervals (CIs) were used to measure the strength of association. The Mantel–Haenszel (MH) test was performed for the combined analysis of the data from both populations. In view of the multiple comparisons, the correction factor n (m − 1) (n loci with m alleles each) was applied to correct the significance level. These analyses were performed using SPSS software (version 15.0). The pairwise linkage disequilibrium (LD) index (Lewontin’s D' and r 2) calculation was performed using the Arlequin package. The haplotype analysis was performed using PLINK software (version 1.07) (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al. 2007).
Results
Population characteristics
A total of 441 laboratory-confirmed patients with EV71 infection and 958 controls were included in the study, including 253 cases and 550 controls from northern China, and 188 cases and 408 controls from southern China. In both populations, controls were comparable with cases with regard to mean age and sex distribution. A total of 150 (59.3 %) patients of northern origin and 151 (80.3 %) cases of southern origin were determined as complicated patients. The characteristics of selected subjects are shown in Table 1.
Table 1.
Characteristics of patients with EV71 infection and control subjects
| Variables | Cases | Controls | P value |
|---|---|---|---|
| No. (%) | No. (%) | ||
| All | (n = 441) | (n = 958) | |
| Northern population | (n = 253) | (n = 550) | |
| Age (months), mean (SD) | 25.3 (13.4) | 27.2 (18.6) | 0.15a |
| Male, no. (%) | 164 (64.8) | 341 (62.0) | 0.44b |
| Clinical diagnosis, no. (%) | |||
| Uncomplicated HFMD | 103 (40.7) | ||
| Complicated cases | 150 (59.3) | ||
| Southern population | (n = 188) | (n = 408) | |
| Age (months), mean (SD) | 33.0 (15.3) | 34.4 (11.2) | 0.19a |
| Male, no. (%) | 118 (62.8) | 238 (58.3) | 0.31b |
| Clinical diagnosis, no. (%) | |||
| Uncomplicated HFMD | 37 (19.7) | ||
| Complicated cases | 151 (80.3) | ||
SD standard deviation
a t test, two-sided
b χ 2 test, two-sided
Individual polymorphism and risk of EV71 infection
The genotyping results were presented in Table 2. The observed genotype frequencies of the C-123A and G-88T polymorphisms were in Hardy–Weinberg equilibrium (all P > 0.05, data not shown). The -123A-positive genotypes were significantly associated with decreased susceptibility to EV71 infection either in the northern population (OR, 0.56; 95 % CI, 0.40–0.81; P = 0.002) or in the southern population (OR, 0.59; 95 % CI, 0.39–0.90; P = 0.01) (Table 2). Similarly, the -88T-positive genotypes were also significantly associated with decreased susceptibility to EV71 infection with an OR of 0.64 (95 % CI, 0.47–0.87; P = 0.004) in the northern population and 0.67 (95 % CI, 0.47–0.95; P = 0.02) in the southern population (Table 2).
Table 2.
The genotype frequencies of MxA polymorphisms in patients with EV71 infection and control subjects
| SNPs and genotypes | Controls | Cases | Crudea | Adjustedb | ||
|---|---|---|---|---|---|---|
| No. (%) | No. (%) | OR (95 % CI) | P value | OR (95 % CI) | P value | |
| Northern population | (n = 550) | (n = 253) | ||||
| C-123A | ||||||
| CC | 379 (68.9) | 197 (79.8) | Reference | Reference | ||
| CA | 161 (29.3) | 46 (18.6) | 0.55 (0.38–0.80) | 0.001 | 0.55 (0.38–0.80) | 0.002 |
| AA | 10 (1.8) | 4 (1.6) | 0.77 (0.24–2.49) | 0.78 | 0.79 (0.24–2.55) | 0.69 |
| CA + AA | 171 (31.1) | 50 (20.2) | 0.56 (0.40–0.81) | 0.002 | 0.56 (0.39–0.81) | 0.002 |
| G-88T | ||||||
| GG | 280 (50.9) | 153 (61.9) | Reference | Reference | ||
| GT | 231 (42.0) | 78 (31.6) | 0.62 (0.45–0.85) | 0.003 | 0.62 (0.46–0.86) | 0.004 |
| TT | 39 (7.1) | 16 (6.5) | 0.75 (0.41–1.39) | 0.36 | 0.76 (0.41–1.41) | 0.39 |
| GT + TT | 270 (49.1) | 94 (38.1) | 0.64 (0.47–0.87) | 0.004 | 0.64 (0.47–0.88) | 0.005 |
| Southern population | (n = 408) | (n = 188) | ||||
| C-123A | ||||||
| CC | 286 (70.6) | 151 (80.3) | Reference | Reference | ||
| CA | 109 (26.9) | 34 (18.1) | 0.59(0.39–0.90) | 0.02 | 0.58 (0.38–0.90) | 0.02 |
| AA | 10 (2.5) | 3 (1.6) | 0.57 (0.15–2.10) | 0.56 | 0.57 (0.15–2.10) | 0.40 |
| CA + AA | 119 (29.4) | 37 (19.7) | 0.59 (0.39–0.90) | 0.01 | 0.58 (0.38–0.89) | 0.01 |
| G-88T | ||||||
| GG | 194 (47.9) | 109 (58.0) | Reference | Reference | ||
| GT | 183 (45.2) | 74 (39.4) | 0.72 (0.50–1.03) | 0.07 | 0.72 (0.50–1.03) | 0.07 |
| TT | 28 (6.9) | 5 (2.7) | 0.32 (0.12–0.85) | 0.02 | 0.33 (0.12–0.87) | 0.03 |
| GT + TT | 211 (52.1) | 79 (42.0) | 0.67 (0.47–0.95) | 0.02 | 0.67 (0.47–0.95) | 0.02 |
| Combined | (n = 958) | (n = 441) | ||||
| C-123A | ||||||
| CC | 665 (69.6) | 348 (80.0) | Reference | Reference | ||
| CA | 270 (28.3) | 80 (18.4) | 0.57 (0.43–0.75)c | <0.001c | 0.56 (0.43–0.75) | <0.001 |
| AA | 20 (2.1) | 7 (1.6) | 0.67 (0.28–1.60)c | 0.37c | 0.68 (0.29–1.63) | 0.39 |
| CA + AA | 290 (30.4) | 87 (20.0) | 0.57 (0.44–0.75)c | <0.001c | 0.57 (0.44–0.75) | <0.001 |
| G-88T | ||||||
| GG | 474 (49.6) | 262 (60.2) | Reference | Reference | ||
| GT | 414 (43.4) | 152 (34.9) | 0.66(0.52–0.84)c | 0.001c | 0.67 (0.53–0.85) | 0.001 |
| TT | 67 (7.0) | 21 (4.8) | 0.57 (0.34–0.95)c | 0.03c | 0.58 (0.34–0.96) | 0.04 |
| GT + TT | 481 (50.4) | 173 (39.8) | 0.65 (0.52–0.82)c | <0.001c | 0.66 (0.52–0.83) | <0.001 |
The number of samples genotyped varies because of genotyping failure for some individuals
OR odds ratio, CI confidence interval
aORs and P values were calculated by logistic regression analysis
bORs and P values were calculated by multivariate logistic regression, adjusted for age (months) and sex
cORs and P values were calculated by Mantel–Haenszel (MH) test
Then multivariate logistic regression analyses were performed by adjusting for age and sex. For the C-123A polymorphism, subjects bearing at least one -123A allele had a decreased susceptibility to EV71 infection compared with those without -123A allele in both independent populations (in the northern population: adjusted OR, 0.56; 95 % CI, 0.39–0.81; P = 0.002; in the southern population: adjusted OR, 0.58; 95 % CI, 0.38–0.89; P = 0.01) (Table 2). For the G-88T polymorphism, subjects bearing at least one -88T allele had a decreased susceptibility to EV71 infection compared with those without -88T allele in both independent populations (in the northern population: adjusted OR, 0.64; 95 % CI, 0.47–0.88; P = 0.005; in the southern population: adjusted OR, 0.67; 95 % CI, 0.47–0.95; P = 0.02) (Table 2). The associations remained significant after correction for multiple comparisons.
When northern and southern population were combined, both the -123A-positive and -88T-positive genotypes were significantly associated with decreased susceptibility to EV71 infection, for the -123A-positive genotype (MH population-weighted OR, 0.57; 95 % CI, 0.44–0.75; P < 0.001), for the -88T-positive genotype (MH population-weighted OR, 0.65; 95 % CI, 0.52–0.82; P < 0.001) (Table 2). There was no significant heterogeneity for both the -123A-positive and -88T-positive genotypes associated ORs among the study populations (Breslow-Day P = 0.87 for C-123A; Breslow-Day P = 0.85 for G-88T).
The effects of C-123A and G-88T genotypes on clinical severity of EV71 infection were also assessed. In both populations, there was no association between G-88T polymorphism and severity of EV71 infection. For C-123A polymorphism, no association was observed with severity of EV71 infection in the southern population. While in the northern population, subjects bearing at least one -123A allele had a decreased susceptibility to EV71 infection compared with those without -123A allele, (adjusted OR = 0.36, 95 % CI = 0.17–0.76, P = 0.007) (Table 3).
Table 3.
The genotype frequencies of MxA polymorphisms in uncomplicated patients and complicated patients
| SNPs and genotypes | Uncomplicated cases | Complicated cases | Crudea | Adjustedb | ||
|---|---|---|---|---|---|---|
| No. (%) | No. (%) | OR (95 % CI) | P value | OR (95 % CI) | P value | |
| Northern population | (n = 103) | (n = 150) | ||||
| C-123A | ||||||
| CC | 72 (71.3) | 125 (85.6) | Reference | Reference | ||
| CA | 26 (25.7) | 20 (13.7) | 0.44 (0.23–0.858) | 0.01 | 0.36 (0.17–0.78) | 0.01 |
| AA | 3 (3.0) | 1 (0.7) | 0.19 (0.02–1.88) | 0.15 | 0.28 (0.02–4.37) | 0.37 |
| CA + AA | 29 (28.7) | 21 (14.4) | 0.42 (0.22–0.79) | 0.006 | 0.36 (0.17–0.76) | 0.007 |
| G-88T | ||||||
| GG | 56 (55.5) | 97 (66.4) | Reference | Reference | ||
| GT | 35 (34.6) | 43 (29.4) | 0.71 (0.41–1.24) | 0.22 | 0.63 (0.33–1.22) | 0.17 |
| TT | 10 (9.9) | 6 (4.1) | 0.35 (0.12–1.00) | 0.06 | 0.51 (0.15–1.75) | 0.29 |
| GT + TT | 45 (44.5) | 49 (33.6) | 0.63 (0.37–1.06) | 0.08 | 0.61 (0.33–1.13) | 0.12 |
| Southern population | (n = 37) | (n = 151) | ||||
| C-123A | ||||||
| CC | 33 (89.2) | 118 (78.2) | Reference | Reference | ||
| CA | 4 (10.8) | 30 (19.9) | 2.10 (0.70–6.38) | 0.24 | 2.07 (0.68–6.31) | 0.20 |
| AA | 0 (0) | 3 (2.0) | NA | NA | NA | NA |
| CA + AA | 4 (10.8) | 33 (21.9) | 2.31 (0.76–6.98) | 0.17 | 2.30 (0.76–6.97) | 0.14 |
| G-88T | ||||||
| GG | 21 (56.8) | 88 (58.3) | Reference | Reference | ||
| GT | 15 (40.5) | 59 (39.1) | 0.94 (0.45–1.97) | 0.87 | 0.94 (0.45–1.97) | 0.86 |
| TT | 1 (2.7) | 4 (2.6) | 0.96 (0.10–8.99) | 1.00 | 1.03 (0.11–9.78) | 0.98 |
| GT + TT | 16 (43.2) | 63 (41.7) | 0.94 (0.45–1.94) | 0.88 | 0.94 (0.45–1.96) | 0.87 |
| Combined | (n = 140) | (n = 301) | ||||
| C-123A | ||||||
| CC | 105 (75.5) | 243 (81.8) | Reference | Reference | ||
| CA | 31 (22.3) | 50 (16.8) | 0.71 (0.42–1.20)c | 0.20c | 0.70 (0.41–1.18) | 0.19 |
| AA | 3 (2.2) | 4 (1.4) | 0.54 (0.11–2.72)c | 0.45c | 0.66 (0.14–3.09) | 0.60 |
| CA + AA | 34 (24.5) | 54 (18.2) | 0.69 (0.42–1.15)c | 0.16c | 0.70 (0.42–1.15) | 0.17 |
| G-88T | ||||||
| GG | 77 (55.4) | 185 (62.3) | Reference | Reference | ||
| GT | 51 (36.7) | 102 (34.3) | 0.79 (0.50–1.22)c | 0.28c | 0.83 (0.54–1.30) | 0.43 |
| TT | 11 (7.9) | 10 (3.4) | 0.42 (0.17–1.06)c | 0.07c | 0.39 (0.16–0.96) | 0.04 |
| GT + TT | 62 (44.6) | 112 (37.7) | 0.72 (0.47–1.10)c | 0.13c | 0.75 (0.49–1.15) | 0.19 |
The number of samples genotyped varies because of genotyping failure for some individuals.
OR odds ratio, CI confidence interval, NA not applicable
aORs and P values were calculated by logistic regression analysis
bORs and P values were calculated by multivariate logistic regression, adjusted for age (months) and sex
cORs and P values were calculated by Mantel–Haenszel (MH) test
Haplotypes and risk of EV71 infection
By LD analysis, the D' values for the MxA C-123A and G-88T polymorphisms in the northern and southern populations were both 1.0, whereas r 2 was 0.48 and 0.44, respectively. Three haplotypes (C- 123–G- 88, C- 123–T- 88 and A- 123–T- 88) were observed based on these two polymorphisms. Consistent with the findings of published studies (Ching et al. 2010; Weinstock-Guttman et al. 2007), the A- 123–G- 88 haplotype was absent in our two populations. The overall haplotype analysis gave a P value of 0.01 in the northern and 0.02 in the southern population (Table 4). Compared with all other haplotypes, the A- 123–T- 88 was associated with a significantly lower risk to EV71 infection in the two independent populations (in the northern population: OR, 0.62; 95 % CI, 0.44–0.85; P = 0.003; in the southern population: OR, 0.63; 95 % CI, 0.43–0.92; P = 0.01) as well as in the combined population (OR, 0.62; 95 % CI, 0.48–0.80; P < 0.001) (Table 4). There was a significantly association between the MxA A- 123–T- 88 haplotype and severity of EV71 infection in the northern population (OR, 0.44; 95 % CI, 0.25–0.79; P = 0.005), which however failed to be detected in the southern population (Table 4).
Table 4.
Haplotypes for MxA gene and their associations with risk to EV71 infection and disease severity
| Haplotypes | Frequency | OR (95 % CI) | P value | Frequency | OR (95 % CI) | P value | ||
|---|---|---|---|---|---|---|---|---|
| Control | Case | Uncomplicated case | Complicated case | |||||
| Northern population | ||||||||
| Globala | 0.01 | 0.01 | ||||||
| Haplotype-specificb | ||||||||
| C-123G-88 | 0.72 | 0.78 | 1.37 (1.06–1.75) | 0.01 | 0.73 | 0.81 | 1.56 (1.03–2.36) | 0.04 |
| A-123T-88 | 0.16 | 0.11 | 0.62 (0.44–0.85) | 0.003 | 0.16 | 0.08 | 0.44 (0.25–0.79) | 0.005 |
| C-123T-88 | 0.12 | 0.11 | 0.97 (0.71–1.34) | 0.87 | 0.11 | 0.11 | 0.99 (0.61–1.62) | 0.98 |
| Southern population | ||||||||
| Globala | 0.02 | 0.08 | ||||||
| Haplotype-specificb | ||||||||
| C-123G-88 | 0.70 | 0.78 | 1.51 (1.12–2.05) | 0.006 | 0.77 | 0.78 | 1.05 (0.55–2.02) | 0.88 |
| A-123T-88 | 0.16 | 0.10 | 0.63 (0.43–0.92) | 0.01 | 0.05 | 0.12 | 2.31 (0.80–6.64) | 0.09 |
| C-123T-88 | 0.14 | 0.12 | 0.84 (0.58–1.23) | 0.36 | 0.18 | 0.10 | 0.48 (0.22–1.04) | 0.07 |
| Combined | ||||||||
| Globala | <0.001 | 0.18 | ||||||
| Haplotype-specificb | ||||||||
| C-123G-88 | 0.71 | 0.78 | 1.42 (1.17–1.73) | <0.001 | 0.74 | 0.79 | 1.37 (0.98–1.92) | 0.07 |
| A-123T-88 | 0.16 | 0.11 | 0.62 (0.48–0.80) | <0.001 | 0.13 | 0.10 | 0.73 (0.47–1.13) | 0.16 |
| C-123T-88 | 0.12 | 0.11 | 0.92 (0.72–1.17) | 0.48 | 0.13 | 0.11 | 0.83 (0.55–1.24) | 0.37 |
OR odds ratio, CI confidence interval
aThe global P value indicates the overall significance of the association of MxA C-123A and G-88T haplotype, determined using the omnibus haplotype test of association
bIn haplotype-specific analyses, the ORs with 95 % CI was determined by testing each haplotype against all others
Quantitative real-time RT-PCR for MxA expression
As shown in Fig. 2, the relative MxA mRNA level from individuals with -123A allele (AA + AC genotypes) was significantly higher than that from individuals with -123CC genotype in both northern and southern populations (t test; P < 0.05). With regard to the G-88T polymorphism, subjects with the -88T allele (TT + GT genotypes) appeared to express higher MxA mRNA than those with the -88GG genotype in both northern and southern populations (t test; P < 0.05). In addition, those subjects that are homozygous for the major C-123A and G-88T haplotype (CC/GG) had lower levels of MxA mRNA than subjects that are heterozygous (CA/GT) in both populations (t test; P < 0.05).
Fig. 2.
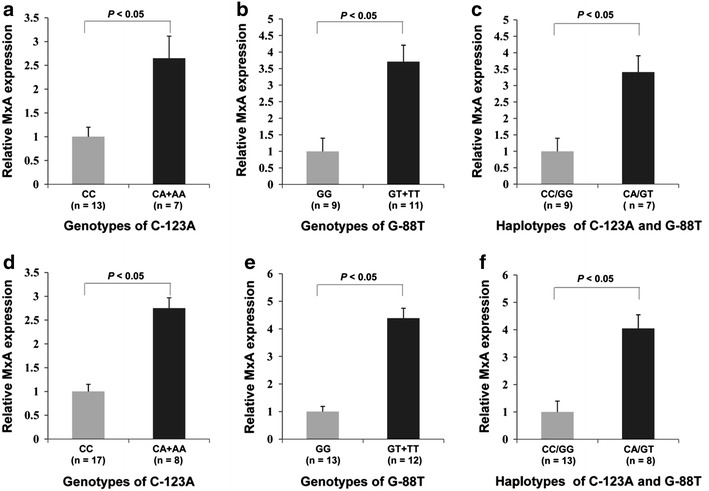
Quantitative real-time RT-PCR for MxA in healthy children. Expression of MxA for three different genotypes of C-123A and G-88T was measured in RNA from 45 healthy children (20 from the northern population and 25 from the southern population) stimulated with IFNβ1a. Normalization for mRNA quantity was performed with human GAPDH control primers for each sample and final abundance figures adjusted to yield an arbitrary value of 1 for -123CC or -88GG genotype. Columns mean from triplicate measurements; the vertical bars indicate the standard deviation (SD). a Correlation of MxA mRNA expression with C-123A genotypes in the northern population. Compared to the CC carriers, the A allele carriers had a markedly elevated MxA transcription (P < 0.05; t test). b Correlation of MxA mRNA expression with G-88T genotypes in the northern population. Compared to the GG carriers, the T allele carriers had a markedly elevated MxA transcription (P < 0.05; t test). c Correlation of MxA mRNA expression with MxA haplotypes in the northern population. The subjects that are homozygous for the major C-123A and G-88T haplotype (CC/GG) had lower levels of MxA mRNA than subjects that are heterozygous (CA/GT) (t test; P < 0.05). d Correlation of MxA mRNA expression with C-123A genotypes in the southern population. Compared to the CC carriers, the A allele carriers had a markedly elevated MxA transcription (P < 0.05; t test). e Correlation of MxA mRNA expression with G-88T genotypes in the southern population. Compared to the GG carriers, the T allele carriers had a markedly elevated MxA transcription (P < 0.05; t test). f Correlation of MxA mRNA expression with MxA haplotypes in the southern population. The subjects that are homozygous for the major C-123A and G-88T haplotype (CC/GG) had lower levels of MxA mRNA than subjects that are heterozygous (CA/GT) (t test; P < 0.05)
The relative MxA mRNA level was also compared between uncomplicated and complicated cases (Fig. 3). There were no significant differences in the relative MxA mRNA level between uncomplicated and complicated cases in both populations (t test; P > 0.05).
Fig. 3.
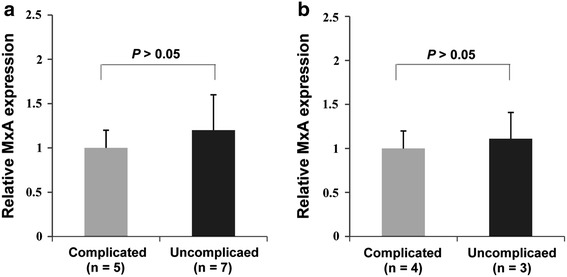
Quantitative real-time RT-PCR for MxA in patients with EV71 infection. Expression of MxA was measured in RNA from 19 patients with EV71 infection (12 from the northern population and 7 from the southern population). Normalization for mRNA quantity was performed with human GAPDH control primers for each sample and final abundance figures adjusted to yield an arbitrary value of 1 for complicated cases. Columns mean from triplicate measurements; the vertical bars indicate the standard deviation (SD). a MxA mRNA expression in patients with EV71 infection in the northern population. Compared to the complicated cases, the uncomplicated cases had a comparable MxA transcription level (P > 0.05; t test). b MxA mRNA expression in patients with EV71 infection in the southern population. Compared to the complicated cases, the uncomplicated cases had a comparable MxA transcription level (P > 0.05; t test)
Discussion
In this study, we found that two functional polymorphisms in the promoter of MxA gene, C-123A and G-88T, were significantly associated with decreased susceptibility to EV71 infection in two independent Chinese subpopulations.Haplotype A- 123–T- 88, derived from these two polymorphisms was also shown to be associated with the decreased risk of EV71 infection. Furthermore, consistent with the results of population-based association study, higher MxA messenger RNA levels in IFNβ1a-stimulated PBMCs from the -123A or -88T allele carriers were observed. To the best of our knowledge, this study is the first to report a genetic association between MxA and susceptibility to EV71 infection, confirming the initial hypothesis that MxA may play a role in the pathogenesis of this disorder.
The genetic association between MxA polymorphism and EV71 infection is biologically plausible. MxA is an antiviral protein induced by type I IFNs that can inhibit viral replication. Following stimulation with type I IFNs, MxA protein accumulates to high levels in the cytoplasm on intracellular membranes. At the point of infection, MxA are released and bind viral nucleocapsids or other viral components, to trap and then degrade them (Sadler and Williams 2008). With regard to EV71, it has been shown that the superior anti-EV71 effect of type I IFN was caused by a strong induction of the downstream antiviral effectors, especially MxA (Sadler and Williams 2008). In addition, it has recently been reported that the antiviral activity of MxA extended to coxsackieviruses B virus, a member of the enterovirus family (Chieux et al. 2001). Thus, it is highly possible that MxA can bind to the components of EV71 and exert its role as an antivirus, although additional studies are needed to clarify this possibility.
The MxA C-123A and G-88T polymorphisms are both functional polymorphisms. Previous in vitro studies showed that -123A and -88T alleles provided stronger binding affinity to nuclear proteins than the wild-type alleles (Ching et al. 2010). Furthermore, the C-123A and G-88T polymorphisms are in strong LD and the A- 123–T- 88 haplotype displays higher promoter activity compared with the C- 123–G- 88 haplotype in luciferase reporter assay (Furuyama et al. 2006; Hijikata et al. 2001; Torisu et al. 2004). In the present study, we also detected higher MxA transcription in IFNβ1a-stimulated PBMCs from the -123A or -88T allele carriers. Given the well-characterized antiviral role of MxA, one might expect that individuals who carry the -123A and/or -88T alleles, and thus have increased expression of MxA, may be at a lower susceptibility to EV71 infection.
In this study, the association with risk of EV71 infection was stronger for the C-123A polymorphism than the G-88T polymorphism. This finding is biologically rational. Although administration of type I IFN can inhibit EV71 replication in vitro and in mouse model, a recent study found that EV71 inhibits the expression of IFN-β in infected cells (Lei et al. 2010), which suggests that the virus evades the antiviral activity of IFN-β. As the C-123A polymorphism affects basal MxA expression without IFN-α and IFN-β induction, whereas the G-88T polymorphism influenced MxA expression after IFN-β stimulation, the C-123A SNP probably exerts greater effect on basal MxA expression in innate immune response against EV71, in which endogenous IFN induction is suppressed. In addition, we found markedly elevated MxA transcription in response to IFNβ1a stimulation for genotypes of the G-88T polymorphism, while the response is not so marked for the C-123A polymorphism. Our results are also consistent with previous findings that the C-123A polymorphism plays a more important role in modifying basal MxA expression, and contributes more significantly to the innate immune response against SARS (Ching et al. 2010).
In this study, we observe associations between MxA C-123A polymorphism and A- 123–T- 88 haplotype with disease severity only in northern population. This result may be due to the limited sample size of patients with uncomplicated cases (n = 37) in the southern population in our study. However, no association between C-123A polymorphism and severity of EV71 infection was observed when northern and southern populations were combined for analysis. Moreover, there were no significant differences in the relative MxA mRNA level between uncomplicated and complicated cases. Therefore, our results indicate that MxA C-123A polymorphism might not play a role in disease severity of EV71 infection. Still, this result warrants further confirmation in future studies.
To our knowledge, only three genetic association studies have addressed susceptibility genes that related to EV71 infection (Chang et al. 2008; Yang et al. 2001, 2012). Some of the results, however, could not be replicated in subsequent studies in other populations. The inconsistent results could be attributable to the small sample size and/or inadequate adjustment for confounding factors such as age and sex. Age and sex are well-known confounding factors for EV71 infection, because individuals aged 1–4 years are associated with a higher risk (Chan et al. 2003; Mao et al. 2010), and the proportion of male was higher in patients with EV71 infection (Chen et al. 2007). This tendency was also identified in our case patients. In the present study, the association was derived from two different subpopulations of large sample size, with potential confounding factors (sex and age) adjusted. Our study, therefore, could probably pose as one of the largest case–control studies demonstrating valid and replicable association of EV71 infection.
Recently, several association studies have shown that the MxA gene was related to the susceptibility to various specific infections (Cao et al. 2009; Ching et al. 2010; Hamano et al. 2005; He et al. 2006; Hijikata et al. 2000). Some of the results, however, could not be replicated in subsequent studies because of multiple factors, such as the different ethnicities of study populations and/or different pathogenesis of the studied diseases. Although the highly significant association between MxA and susceptibility to EV71 derived from a biologically based a priori hypothesis, our findings should be independently verified in populations of different ancestry, such as white subjects and African subjects.
In conclusion, our study shows an association between the MxA polymorphisms and decreased susceptibility to EV71 infection in two Chinese subpopulations. These findings may provide support for the importance of MxA in the pathogenesis of EV71 infection. If confirmed by other studies, knowledge of genetic factors contributing to the pathogenesis of EV71 infection as presented here would be important for the assessment of host susceptibility to EV71 infection and other infectious diseases, especially those sharing a mode of action similar to that of EV71 infection.
Acknowledgments
We thank all the tested individuals, their families, and collaborating clinicians for their participation. This work was supported by grants from the China Mega-Project on Infectious Disease Prevention (No. 2013ZX10004202-002), National Science Fund for Young Scholars (No. 81222037), National Natural Science Foundation (No. 81202253) and Shandong Scientific and Technical Supporting Program (No. 2009GG10002055).
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Wei Liu, Phone: +86-10-63896082, FAX: +86-10-63896082, Email: liu_weis@sohu.com.
Wuchun Cao, Phone: +86-10-63896082, FAX: +86-10-63896082, Email: caowc@bmi.ac.cn.
References
- Cao B, Liu X, Hou F, Li W, Han Z, Zhang Q, Dai Y, Xu C, Qi H. The haplotype of the MxA gene promoter is associated with hepatitis B virus infection in a Chinese population. Liver Int. 2009;29:1383–1388. doi: 10.1111/j.1478-3231.2009.02053.x. [DOI] [PubMed] [Google Scholar]
- Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis. 2003;9:78–85. doi: 10.3201/eid1301.020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LY, Chang IS, Chen WJ, Huang YC, Chen GW, Shih SR, Juang JL, Shih HM, Hsiung CA, Lin TY, Huang LM. HLA-A33 is associated with susceptibility to enterovirus 71 infection. Pediatrics. 2008;122:1271–1276. doi: 10.1542/peds.2007-3735. [DOI] [PubMed] [Google Scholar]
- Chen SC, Chang HL, Yan TR, Cheng YT, Chen KT. An eight-year study of epidemiologic features of enterovirus 71 infection in Taiwan. Am J Trop Med Hyg. 2007;77:188–191. [PubMed] [Google Scholar]
- Chen SP, Huang YC, Li WC, Chiu CH, Huang CG, Tsao KC, Lin TY. Comparison of clinical features between coxsackievirus A2 and enterovirus 71 during the enterovirus outbreak in Taiwan, 2008: a children’s hospital experience. J Microbiol Immunol Infect. 2010;43:99–104. doi: 10.1016/S1684-1182(10)60016-3. [DOI] [PubMed] [Google Scholar]
- Chieux V, Chehadeh W, Harvey J, Haller O, Wattre P, Hober D. Inhibition of coxsackievirus B4 replication in stably transfected cells expressing human MxA protein. Virology. 2001;283:84–92. doi: 10.1006/viro.2001.0877. [DOI] [PubMed] [Google Scholar]
- Ching JC, Chan KY, Lee EH, Xu MS, Ting CK, So TM, Sham PC, Leung GM, Peiris JS, Khoo US. Significance of the myxovirus resistance A (MxA) gene −123C >a single-nucleotide polymorphism in suppressed interferon beta induction of severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2010;201:1899–1908. doi: 10.1086/652799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Arcas N, Blanco A, Gaitan MJ, Nyqvist M, Alonso A, Reyes-Engel A. Differential transcriptional expresion of the polymorphic myxovirus resistance protein A in response to interferon-alpha treatment. Pharmacogenetics. 2004;14:189–193. doi: 10.1097/00008571-200403000-00007. [DOI] [PubMed] [Google Scholar]
- Furuyama H, Chiba S, Okabayashi T, Yokota S, Nonaka M, Imai T, Fujii N, Matsumoto H. Single nucleotide polymorphisms and functional analysis of MxA promoter region in multiple sclerosis. J Neurol Sci. 2006;249:153–157. doi: 10.1016/j.jns.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Hamano E, Hijikata M, Itoyama S, Quy T, Phi NC, Long HT, Ha LD, Ban VV, Matsushita I, Yanai H, Kirikae F, Kirikae T, Kuratsuji T, Sasazuki T, Keicho N. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Feng D, de Vlas SJ, Wang H, Fontanet A, Zhang P, Plancoulaine S, Tang F, Zhan L, Yang H, Wang T, Richardus JH, Habbema JD, Cao W. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect Dis. 2006;6:106. doi: 10.1186/1471-2334-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Ohta Y, Mishiro S. Identification of a single nucleotide polymorphism in the MxA gene promoter (G/T at nt −88) correlated with the response of hepatitis C patients to interferon. Intervirology. 2000;43:124–127. doi: 10.1159/000025035. [DOI] [PubMed] [Google Scholar]
- Hijikata M, Mishiro S, Miyamoto C, Furuichi Y, Hashimoto M, Ohta Y. Genetic polymorphism of the MxA gene promoter and interferon responsiveness of hepatitis C patients: revisited by analyzing two SNP sites (−123 and −88) in vivo and in vitro. Intervirology. 2001;44:379–382. doi: 10.1159/000050075. [DOI] [PubMed] [Google Scholar]
- Kochs G, Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci USA. 1999;96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, He B, Wang J. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J Virol. 2010;84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Lee YP, Wang YF, Lei HY, Liu CC, Wang SM, Su IJ, Wang JR, Yeh TM, Chen SH, Yu CK. Type I interferons protect mice against enterovirus 71 infection. J Gen Virol. 2005;86:3263–3269. doi: 10.1099/vir.0.81195-0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mao LX, Wu B, Bao WX, Han FA, Xu L, Ge QJ, Yang J, Yuan ZH, Miao CH, Huang XX, Zhang C, Xu H. Epidemiology of hand, foot, and mouth disease and genotype characterization of Enterovirus 71 in Jiangsu, China. J Clin Virol. 2010;49:100–104. doi: 10.1016/j.jcv.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Pavlovic J, Arzet HA, Hefti HP, Frese M, Rost D, Ernst B, Kolb E, Staeheli P, Haller O. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J Virol. 1995;69:4506–4510. doi: 10.1128/jvi.69.7.4506-4510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Hochkeppel HK, Horisberger MA, Hovanessian A, Haller O. MxA gene expression after live virus vaccination: a sensitive marker for endogenous type I interferon. J Infect Dis. 1994;169:807–813. doi: 10.1093/infdis/169.4.807. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- Torisu H, Kusuhara K, Kira R, Bassuny WM, Sakai Y, Sanefuji M, Takemoto M, Hara T. Functional MxA promoter polymorphism associated with subacute sclerosing panencephalitis. Neurology. 2004;62:457–460. doi: 10.1212/01.WNL.0000106940.95749.8E. [DOI] [PubMed] [Google Scholar]
- Verstrepen WA, Bruynseels P, Mertens AH. Evaluation of a rapid real-time RT-PCR assay for detection of enterovirus RNA in cerebrospinal fluid specimens. J Clin Virol. 2002;25(Suppl 1):S39–S43. doi: 10.1016/S1386-6532(02)00032-X. [DOI] [PubMed] [Google Scholar]
- von Wussow P, Jakschies D, Block B, Tschechne B, Schedel I, Horisberger MA, Hochkeppel HK, Deicher H. The interferon-induced Mx-homologous protein in people with symptomatic HIV-1 infection. AIDS. 1990;4:119–124. doi: 10.1097/00002030-199002000-00004. [DOI] [PubMed] [Google Scholar]
- Weinstock-Guttman B, Tamano-Blanco M, Bhasi K, Zivadinov R, Ramanathan M. Pharmacogenetics of MXA SNPs in interferon-beta treated multiple sclerosis patients. J Neuroimmunol. 2007;182:236–239. doi: 10.1016/j.jneuroim.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Xie P, Tang Y, Shen S, Wang Y, Xing G, Yin Y, He F, Zhang L. Smurf1 ubiquitin ligase targets Kruppel-like factor KLF2 for ubiquitination and degradation in human lung cancer H1299 cells. Biochem Biophys Res Commun. 2011;407:254–259. doi: 10.1016/j.bbrc.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhao N, Su NL, Sun JL, Lv TG, Chen ZB. Association of interleukin 10 and interferon gamma gene polymorphisms with enterovirus 71 encephalitis in patients with hand, foot and mouth disease. Scand J Infect Dis. 2012 doi: 10.3109/00365548.2011.649490. [DOI] [PubMed] [Google Scholar]
- Yang KD, Yang MY, Li CC, Lin SF, Chong MC, Wang CL, Chen RF, Lin TY. Altered cellular but not humoral reactions in children with complicated enterovirus 71 infections in Taiwan. J Infect Dis. 2001;183:850–856. doi: 10.1086/319255. [DOI] [PubMed] [Google Scholar]
- Yi L, He Y, Chen Y, Kung HF, He ML. Potent inhibition of human enterovirus 71 replication by type I interferon subtypes. Antivir Ther. 2011;16:51–58. doi: 10.3851/IMP1720. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li X, Wu Z, Lin F, Zhou H. The p73 G4C14-to-A4T14 polymorphism is associated with risk of lung cancer in the Han nationality of North China. Mol Carcinog. 2012 doi: 10.1002/mc.21869. [DOI] [PubMed] [Google Scholar]


