Abstract
NRAS is a commonly mutated oncogene in melanoma and therapeutic targeting of NRAS has proven difficult. We characterized the expression and phenotypic functions of 5 recently discovered splice isoforms of NRAS in melanoma. Canonical NRAS (isoform-1) was expressed to the highest degree and its expression was significantly increased in melanoma metastases compared to primary lesions. Isoform-5 expression in metastases showed a significant, positive correlation with survival and tumors over-expressing isoform-5 had significantly decreased growth in a xenograft model. In contrast, over-expression of any isoform resulted in enhanced proliferation, and invasiveness was increased with over-expression of isoform-1 or isoform-2. Downstream signaling analysis indicated that the isoforms signaled differentially through the MAPK and PI3K pathways and A375 cells over-expressing isoform-2 or isoform-5 showed resistance to vemurafenib treatment in vitro. The NRAS isoforms appear to play varying roles in melanoma phenotype and could potentially serve as biomarkers for therapeutic response and disease prognosis.
Keywords: Melanoma, NRAS, isoforms
INTRODUCTION
Melanoma is a malignancy that arises from the melanocytes, or pigment producing cells in the skin [1]. It is the most deadly form of skin cancer and the incidence is continuing to rise, with an estimated 87,000 new cases being diagnosed in the United States in 2017 [2, 3]. While surgical excision remains the primary treatment modality for early stage disease, treatment options for advanced melanoma are expanding to include the use of immunotherapeutic agents as well as targeted inhibitors of signal transduction, in addition to standard chemotherapy [4]. Despite these advances, nearly 10,000 deaths from melanoma are predicted for this year [2].
Neuroblastoma RAS viral oncogene homolog (NRAS), one of the three RAS genes, encodes a small plasma membrane-associated GTP-binding protein which functions to link signals from receptor tyrosine kinases on the cell surface to nuclear transcription factors [5]. NRAS is the second most commonly mutated oncogene in melanoma; it is altered in 15–20% of cases and was the first melanoma oncogene to be discovered [6, 7]. Despite intensive research efforts, the development of NRAS-specific therapies has proven difficult, and thus current treatment modalities for NRAS-mutated melanoma center on the targeting of downstream signal transduction pathways that drive RAS-mediated transformation [8, 9].
Alternative splicing of genes/transcripts has been shown to be the major mechanism for the generation of proteome diversity from limited genetic material [10]. It is now known that over 90% of human genes undergo alternative splicing and that the wild-type protein is often not the dominant transcript present [11, 12]. It has also been shown that the majority of alternatively transcribed isoforms are functionally distinct and behave more like separate proteins, rather than merely variants of each other [13]. Recently, four previously unknown isoforms of NRAS (isoforms 2–5) in addition to the canonical NRAS (isoform 1) were discovered and were shown to have varying expression patterns, enzymatic activity, and downstream oncogenic effects (Figure 1, Ref. 14). These isoforms result from the introduction of a previously unknown exon (exon 3b, isoform 2), the skipping of exon 3 (isoform 3), the skipping of exons 3 and 4 (isoform 4), or the fusion of the beginning of exon 2 with the end of exon 5 (isoform 5; Fig. 1). The structure of the smallest of the isoforms, isoform 5, was recently characterized by our group, but little is known beyond these initial descriptions [15]. Thus, it is essential to thoroughly examine the functional role of these isoforms in melanoma, as NRAS plays a pivotal role in this malignancy.
Figure 1: Structure of the NRAS isoforms.

Isoform 1 is the canonical NRAS isoform with an open reading frame spanning exons 2 through 5. Isoform 2 contains an additional exon: 3b. Isoforms 3 and 4 are lacking exon 3 or exons 3 and 4, respectively. Isoform 5 contains a fusion of the first 17 codons of exon 2 with 3 codons near the end of exon 5. Depicted are cDNA, mRNA splicing products, and protein lengths of the 5 isoforms. Numbered boxes represent the exons (Adapted from ref. 14).
In the present study, the expression patterns of the five NRAS isoforms in melanoma tumor tissues were characterized. Their phenotypic function in melanoma cells was also evaluated, as well as their implications for the melanoma response to common therapeutics. This information will help to clarify what role these NRAS splice variants play in melanoma, as well as their potential to serve as therapeutic targets or biomarkers in this disease setting.
METHODS
Tumor Tissue and Cell Lines
The A375 human metastatic melanoma cell line was obtained from American Type Cell Culture Collection (ATCC, Manassas, VA). Melanoma tumor tissue samples were obtained from the OSU Wexner Medical Center. All patients provided written informed consent to store and use their tissue for discovery studies according to OSU institutional guidelines under protocols approved by the OSU Institutional Review Board.
Quantification of the NRAS Isoforms From Melanoma Tumor Tissue
Total RNA from cells was isolated using TRIzol reagent (Invitrogen) as per the manufacturer’s recommendations. Isoform-specific Taqman qPCR primers were designed for each isoform. Specificity was confirmed by using RNA from cells after forced isoform expression. Primer sequences used for the qPCR were as followed: NRAS isoform 1 forward, TAACCTCTACAGGGAGCAGAT; NRAS isoform 1 reverse, GTGGGCTTGTTTTGTATCAAC; NRAS isoform 2 forward, CTACAGGCTGGAGTGCAG; NRAS isoform 2 reverse, GTCTTTTACTCGCTTAATCTG; NRAS isoform 2 forward, CCACCATAGAGGGAGCAGA; NRAS isoform 3 reverse, GCTTGTTTTGTATCAACTGTCC; NRAS isoform 4 forward, CACCATAGAGGGTGTTGAAG; NRAS isoform 4 reverse, CACACATGGCAATCCCATAC; NRAS isoform 5 forward, CGTGTGAAATGACTGAGTAC; and NRAS isoform 5 reverse, ATCACCACACATGGCTTTTC. Relative expression was normalized to 18S rRNA as a housekeeping gene.
Analysis of Publicly Available RNA-seq and Clinical Data
Controlled-access raw RNA-seq data from melanoma patient tumors in The Cancer Genome Atlas Skin Cutaneous Melanoma (TCGA SKCM) subset (n=472 tumor samples) was downloaded from the National Cancer Institute Genomics Data Commons Legacy Archive (https://gdc-portal.nci.nih.gov/legacy-archive/). Corresponding clinical and biomolecular data was accessed via the cBioPortal for Cancer Genomics and Firebrowse data portal (http://firebrowse.org) [16, 17]. In order to generate SAM alignment files, the STAR program (version 2.4.2a) was used to align fastq files to a modified version of the GENCODE protein-coding transcript sequences (release 24) human reference genome multi-FASTA file that included the 5 NRAS isoform sequences [18] We used the eXpress program (version 1.5.1) to generate read counts from the SAM files and determine transcript abundances in units of transcripts per million (TPM) and effective counts [19]. Overall and disease-free survival analyses for the TCGA SKCM patients were performed using Prism GraphPad software in order to generate Kaplan-Meier plots and calculate P values using the log-rank test [16, 17].
Forced Overexpression of the NRAS Isoforms
For stable expression, the isoforms were shuttled into a lentiviral expression vector and transduced into the A375 melanoma cell line utilizing the LentiStarter 2.0 kit (SBI). Overexpression was confirmed via qPCR as previously described [14].
Functional Assays
Cell proliferation was assessed using the Cell Titer 96 Aqueous One Solution Cell Proliferation MTS Assay (Promega). Transformation assays were performed using the CytoSelect 96-well Cell Transformation Assay Kit (Soft Agar Colony Formation; Cell Biolabs). Matrigel invasion assays were conducted according to the manufacturer’s instructions (BD Biosciences) and photographs were taken of five 20X fields for each filter on an EVOS XL digital inverted microscope as previously described [20].
Immunoblot Analysis
Cells were collected and lysed in RIPA buffer (Sigma Aldrich, St. Louis, MO) containing protease inhibitor and phosphatase inhibitor cocktails (Thermo Fisher Scientific, Inc., Waltham, MA). Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride filters and probed with the following antibodies: phosphorylated (P)-AKT (S473, 4060S; Cell Signaling), AKT (no. 9272; Cell Signaling), Actin (sc-1616; Santa Cruz), P-ERK (9101S; Cell Signaling), ERK (4695P; Cell Signaling), and anti-rabbit IgG (7074S; Cell Signaling). Following incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies, immune complexes were detected using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc., Waltham, MA). β-actin was used to confirm equal loading.
Proliferation Assay Following Vemurafenib Treatment
The proliferation of melanoma cells treated with vemurafenib was measured as absorbance at 490 nm using the Cell Titer 96 Aqueous One Solution Cell Proliferation MTS Assay (Promega). All assays were performed in triplicate. Cells were also stained with trypan blue (Life Technologies, Carlsbad, CA) and live/dead cell counting was used to confirm proliferation assay results.
In vivo Studies
Female 4–6 week old nude mice (Jackson Laboratories, Bar Harbor, ME) were used for tumor growth studies. 106 A375 melanoma cells were injected subcutaneously in the right flank. Tumors were measured every other day.
Statistical Methods
Statistical significance of differences between groups was analyzed by ANOVA and Student’s t-test, and p ≤ 0.05 was considered to be statistically significant. P values were adjusted for multiple comparisons using the Holm-Bonferroni method. For mouse tumor studies, a linear mixed model was employed to model longitudinal tumor volume for mice under each treatment. Comparisons were done at each time point and averaged across all time points using t-statistics.
RESULTS
NRAS Isoforms Are Expressed In Melanoma Tumor Tissue
To thoroughly understand the spectrum of NRAS isoform expression in melanoma, quantitative PCR was utilized, as antibodies do not exist that are specific for the NRAS isoforms and their similar molecular weights make it challenging to resolve isoforms from the full-length protein. Isoform expression was determined via qPCR in a panel of 21 melanoma tumor tissues, representing metastases to various locations (Supplemental Table 1). Isoform 1 (canonical NRAS) was found to have the highest overall mRNA expression followed by isoforms 3, 5, 4 and 2, respectively (Fig 2A). Publicly available RNA-seq gene expression data from the The Cancer Genome Atlas’s (TCGA) skin cutaneous melanoma subset was also analyzed. The effective counts and transcripts per million (TPM) for each of the five NRAS isoforms across all patient samples were determined (n=472, Figure 2B). Similar patterns of isoform expression were seen in the TCGA cohort, with isoform 1 expressed to the highest degree, followed by the other four splice variants. However, all isoforms were readily detected in these samples.
Figure 2: NRAS isoforms are expressed in melanoma tumor tissues.

(A) Expression analysis by qRT-PCR of NRAS isoforms 1–5 mRNA in 21 melanoma tumor tissues. (B) Transcript abundance in transcripts per million of NRAS isoforms 1–5 from publicly available RNAseq data of 472 skin cutaneous melanoma tumor tissues. (C) Expression analysis by qRT-PCR of NRAS isoforms 1–5 mRNA in 23 additional melanoma tumor tissues representing primary lesions and metastases to various locations. (D) Transcript abundance in transcripts per million of NRAS isoforms 1–5 from publicly available RNAseq data of 472 skin cutaneous melanoma tumor tissues separated into primary or metastatic samples. Bars represent mean ± s.e.m. *** p<0.001.
NRAS Isoform Levels Vary From Primary Lesions to Metastases
Next, a panel of melanoma tissues representing primary lesions (n=3), hepatic metastases (n=4), brain metastases (n=10) and pulmonary metastases (n=6) was evaluated via qPCR for NRAS isoform expression levels (Figure 2C). The isoforms showed distinct patterns of expression; levels of isoforms 1 and 3 increased from primary lesions to all of the metastatic locations, with negligible differences in expression between the various metastatic locations. Isoform 2 levels decreased slightly from primary lesions to metastases, but the differences were minimal. Isoform 4 expression was increased in hepatic and brain metastases, but decreased in pulmonary metastases when compared to primary lesions. Isoform 5 levels increased from primary to metastases, with brain and pulmonary metastases having the highest overall expression. However, likely due to the small sample size, no statistically significant differences between isoform expression levels were observed. When the TCGA samples were separated into primary lesions and metastases, only isoform 1 showed a significant difference in expression, increasing by approximately 10 TPM in metastatic samples versus primaries (p<0.001, Figure 2D).
Elevated Isoform 5 Expression in Melanoma Metastases is Associated With Enhanced Survival
Next, the relationship between NRAS isoform expression and melanoma patient survival was evaluated. When the entire TCGA melanoma cohort was analyzed for survival based on high or low expression of each of the NRAS isoforms, there were no significant differences in survival. However, when metastatic samples alone were stratified based on expression of each of the isoforms, patients with top quartile expression of isoform 5 had significantly increased survival when compared to patients with isoform 5 levels in the bottom quartile (p=0.017, Figure 3). Increased expression of isoform 4 in metastases also showed a trend towards increased survival, though this was not statistically significant (p=0.059). Expression of NRAS isoforms 1, 2, and 3 in melanoma metastases showed no correlations with survival.
Figure 3: Relationship of NRAS isoform levels to survival in melanoma.

(A) Overall survival of the skin cutaneous melanoma patient cohort from the TCGA database separated by top quartile (blue) or bottom quartile (red) isoform expression as measured by RNAseq.
NRAS Isoform Over-expression Leads to Changes in Phenotype and Signaling
To further characterize the effect of isoform expression on the phenotype of melanoma cells the A375 melanoma cell line was transduced utilizing lentiviral vectors containing each of the five NRAS isoforms, and these genetically altered cell lines were analyzed for their proliferative capacity, invasiveness, anchorage-independent growth and downstream signaling (Fig. 4). All of the newly discovered isoforms were successfully over-expressed, as measured by qPCR (Supplemental figure S1). The proliferative ability of the cell lines was significantly altered by over-expression of each of the individual isoforms; over-expression of any of the 5 isoforms resulted in significantly increased proliferation at both the 48 and 72 hour time points when compared to the empty vector control cell line, with isoform 3 over-expression resulting in the highest proliferation at both time points, approximately 2.5-fold higher than the empty vector cell line (Fig. 4A). Interestingly, the cell lines that over-expressed isoform 1 or 2 (A375 iso 1 or 2) also exhibited significantly increased invasive capacity when compared to the other isoforms and the empty vector-transduced control cell line suggesting a more mobile phenotype (Fig. 4B). In a soft agar-based assay, the melanoma cell line over-expressing isoform 5 showed the greatest ability for anchorage-independent growth, while isoform 4 over-expression led to a significant decrease in this growth when compared to the empty vector cell line (p<0.05). All other cell lines showed comparable levels of colony formation in soft agar (Fig. 4C).
Figure 4: Phenotypic characterization of NRAS isoform over-expressing cell lines.

(A) MTS proliferation assay measuring cell proliferation after 48 or 72 hours of A375 cell line transduced with an empty vector control (A375 EV) or A375 transduced with vectors specific for each NRAS isoform (A375 iso 1–5). (B) Quantification of matrigel invasion assay, (C) Visualization of soft agar colony formation assay at day 4, 4x magnification of 96 well plate and quantification of cell growth by fluorescent staining, and (D) Immunoblot probing for p-ERK, p-AKT, total ERK and AKT and β-actin of same panel of A375 cell lines (left), and densitometry analysis for n=3 immunoblots (right). Bars represent three biological replicates, displayed as mean ± s.e.m. * p <0.05, ** p<0.01, *** p< 0.001.
NRAS, when activated, can lead to phosphorylation and subsequent activation of various downstream signal transducers such as ERK and AKT. In order to test the effect of isoform over-expression on the activation of these proteins, lysates of the various isoform over-expressing cell lines were probed for the native and phosphorylated forms of these factors by immunoblot. Interestingly, the A375 cell lines that over-expressed isoforms 1 and 3 exhibited phosphorylation of ERK similar to that of the empty vector-transduced cell line, while over-expression of isoforms 2, 4 and 5 resulted in decreased ERK activation. Evaluation of these same lysates for levels of p-AKT revealed increased phosphorylation of AKT with over-expression of isoforms 2 and 3 and a decreased phosphorylation with over-expression of isoforms 1, 4 and 5, when compared to p-AKT levels in the empty vector transduced cell line (A375 EV).
NRAS Isoform over-expression alters tumor growth in vivo.
To better understand the effects of NRAS isoform over-expression on melanoma tumor growth, athymic nude mice were inoculated with the A375 cell lines that had been engineered to over-express each NRAS isoform or the empty vector control. Tumor growth was monitored for 27 days and no significant differences were seen in tumor growth between the various NRAS isoform over-expressing cell lines as compared to the control cell line, except for the isoform 5 over-expressing cell line which had significantly decreased tumor growth (p<0.05, Fig. 5A). Additionally, tumors composed of cells over-expressing isoform 5 were the smallest at the completion of the study (Fig. 5B–C). In contrast, tumors over-expressing isoform 1 or 4 had similar tumor growth kinetics to the empty vector tumors and resulted in similarly large tumors at the end of the study.
Figure 5: NRAS Isoform over-expression alters tumor growth in vivo.
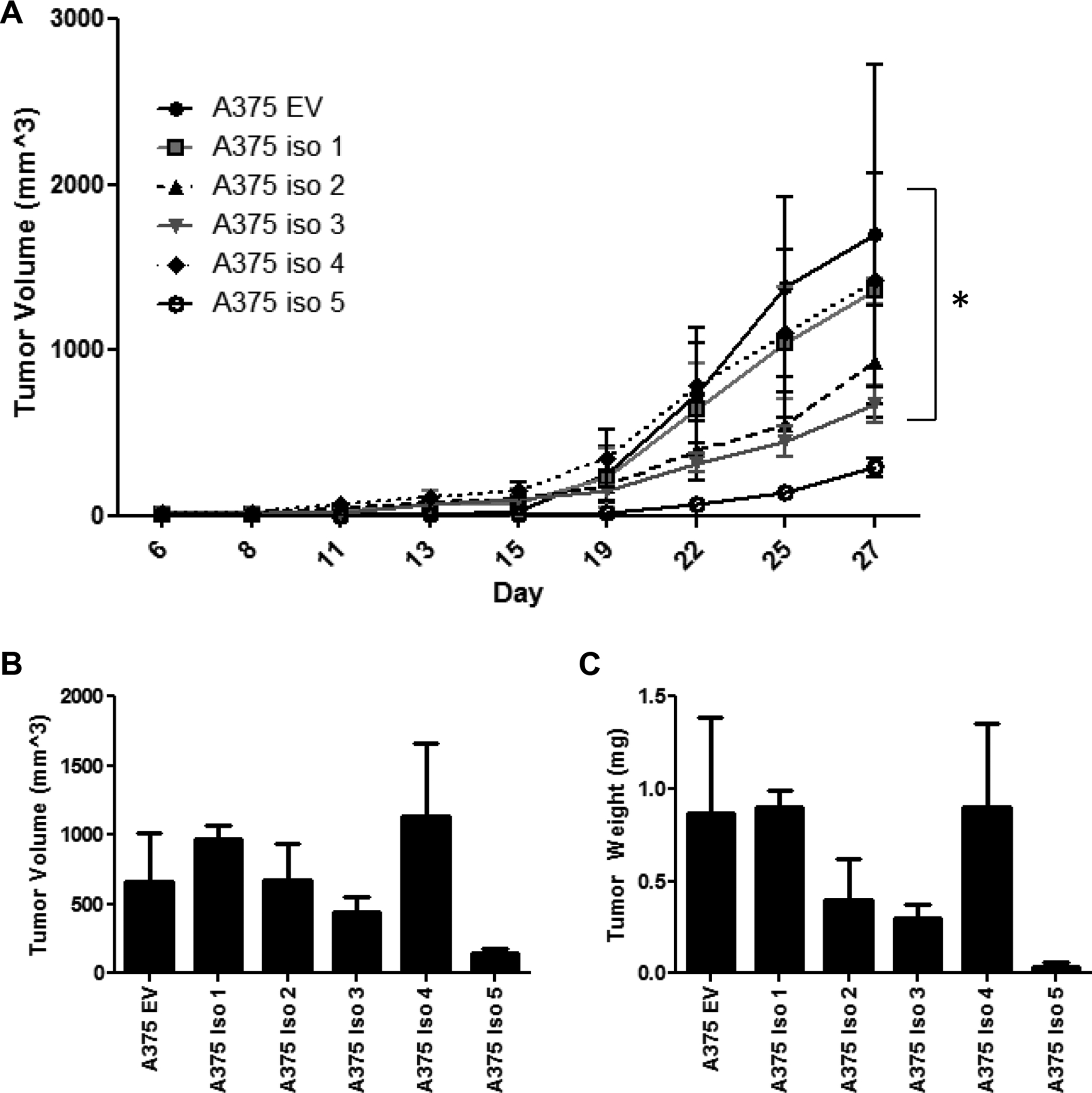
(A) A375 cells that over-express either NRAS isoforms 1–5 or an empty vector control (A375 EV) were injected s.c. into athymic nude mice (1×106 cells). and tumor growth was followed. On day 27, mice were sacrificed and tumors were removed and measured for volume (B) and weight (C). Bars depict mean ± s.e.m.
Over-expression of NRAS Isoform 2 or 5 Enhances Resistance to Vemurafenib In Vitro
The effect of vemurafenib exposure on the behavior of the modified cell lines was next examined (Fig. 6). Notably, the A375 cell lines over-expressing isoforms 2 and 5 demonstrated significantly greater proliferation following 48 hours of vemurafenib treatment compared to controls than did the other cell lines (Fig. 6A). Conversely, isoform over-expression did not appear to alter cell apoptosis following treatment with other common melanoma therapeutics including paclitaxel, dacarbazine, and IFN-α (data not shown). BRAF inhibitor therapies, including vemurafenib and dabrafenib, are indicated for melanoma patients with activating BRAF V600 E/K mutations, which are often mutually exclusive to oncogenic NRAS mutations. Therefore, we determined the proportion of melanoma tumors with high vs. low mRNA expression of each of the NRAS isoforms in BRAF mutant vs. wild type and NRAS mutant vs. wild-type tumors (Fig. 6B). mRNA expression levels of isoforms 2 and 3 (top vs. bottom quartile) demonstrated significant differences in the proportion of BRAF mutant vs. wild-type (wt) tumors (Fisher’s exact test, p= 0.0486, p= 0.0310, respectively), where the top quartile of both isoforms was lower in BRAF-mutant tumors and higher in BRAF-wt tumors. Conversely, a higher proportion of isoforms 1, 2 and 3 were found to be present in NRAS-mutant tumors and lower in NRAS-wt tumors (Fisher’s exact test, p <0.0001, p=0.0057, p=0.0100, respectively). Thus, isoforms 2 and 3 were lower in the presence of the BRAF mutation whereas isoforms 1, 2, and 3 were higher in the presence of the NRAS mutation.
Figure 6: NRAS Isoform expression alters response to vemurafenib treatment.

(A) Proliferation of NRAS isoform over-expressing cell lines in the presence of vemurafenib doses from 0–20 μM was measured by MTS assay after 48 hours of drug exposure, displayed as mean ± s.e.m of three replicate experiments. (B) Proportion of NRAS isoforms in BRAF/NRAS-mutant vs. wild-type (wt) metastatic melanoma patient tumors. The top and bottom quartile of NRAS isoform mRNA expression (RNAseq TCGA data in transcripts per million (TPM)). P values were determined via Fisher’s exact test.
DISCUSSION
NRAS is the second most commonly mutated oncogene in melanoma cancers and attempts to target NRAS therapeutically have proven challenging. This work sought to characterize the expression of recently discovered NRAS isoforms in human melanoma and to understand their oncogenic functions and roles in response to melanoma therapeutics. A summary of this characterization is described in Table 1. All five NRAS isoform transcripts were found to be expressed in melanoma tumor tissue, with canonical NRAS (isoform 1) consistently expressed to the highest degree. NRAS isoform 1 mRNA expression was also significantly increased in metastases compared to primary melanoma lesions. Only isoform 5 showed a significant correlation with survival, as high levels of isoform 5 in melanoma metastases were associated with enhanced survival in these patients. Forced over-expression of each the isoforms led to enhanced proliferation, but invasiveness was only increased with over-expression of isoforms 1 or 2. Notably, over-expression of isoform 4 led to significantly decreased ability for anchorage independent cell growth. Downstream signaling analysis indicated that the isoforms varied in their signaling through the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase pathways, and in vivo growth of A375 cells over-expressing each of the isoforms found that cells over-expressing isoform 5 had significantly decreased tumor growth. Finally, A375 cells over-expressing isoforms 2 or 5 showed marked resistance to vemurafenib treatment in vitro.
Table 1:
Summary of NRAS Isoform Functions in Melanoma
| Isoform | mRNA Structure | Expression | Survival | Phenotype | Signaling | Drug Response |
|---|---|---|---|---|---|---|
| 1 | exons 2, 3, 4, 5 |
|
no effect |
|
|
no effect |
| 2 | exons 2, 3, 3b, 4, 5 |
|
no effect |
|
|
increased vemurafenib resistance |
| 3 | exons 2, 4, 5 |
|
no effect |
|
|
no effect |
| 4 | exons 2, 5 |
|
high expression ~ increased survival in metastases |
|
|
no effect |
| 5 | fusion of beginning of exon 2 with end exon 5 |
|
high expression -> increased survival in metastases |
|
|
increased vemurafenib resistance |
The studies herein indicate that these novel NRAS isoforms have distinct biological activities which may have clinical relevance. In line with the initial discovery publication by Eisfeld et al., we found that each isoform differentially affected melanoma cell oncogenic functions and signaling pathways [14]. Not surprisingly, canonical NRAS isoform 1 was expressed to the highest degree in all melanoma tissues evaluated. Given the known oncogenic functions of NRAS, it was also not surprising that when over-expressed, isoform 1 led to increased proliferative and invasive potential of melanoma cells. However, high levels of isoform 1 in tumor tissue did not correlate with worsened survival and thus an increase in expression of canonical NRAS alone is likely not sufficient to drive disease progression.
Isoforms 4 and 5 showed a potentially protective effect in melanoma metastases, as higher expression of these isoforms correlated with increased survival. Isoform 4 over-expressing cells also had significantly decreased anchorage independent growth in a soft agar assay, while isoform 5 over-expressing cells grew the slowest in vivo. Both of these isoforms appeared to down-regulate MAPK pathway signaling, as indicated by p-ERK, which may explain some of the less oncogenic properties of these isoforms. Interestingly, our previous experiments on the effects of isoform 5 over-expression were conducted in COS-7 monkey fibroblast cells and revealed increased activation of multiple NRAS targets [14]. The differences in downstream signaling seen with isoform 5 over-expression in A375 cells are likely due to differences between these two cell lines. Notably, isoform 5 is made up of only 20 amino acids (first 17 codons of exon 2 with 3 codons toward the end of exon 5) and it was previously shown to be localized to the nucleus with no GTPase activity [14,15]. How it exerts its effects remains unclear and is under investigation by our group.
In this investigation we chose not to stably over-express myc-tagged isoforms, as utilized in the Eisfeld publication, due to concerns that myc-tagging might interfere with the function and signaling of the isoforms [14]. However, it will be important to investigate protein levels of the isoforms in future characterizations.
It is important to note that only isoforms 1 and 2 contain the mutational hotspot codon 61 (exon 3) which is the most frequent point of mutation in melanoma [21]. Thus, it will be essential to characterize these isoforms in the context of this mutation, as well. The proportion of melanoma tumors with high mRNA expression of isoforms 1, 2 and 3 were significantly increased in NRAS-mutant melanoma tumors identified via TCGA database as compared to wild-type tumors. The proportion of tumors with high mRNA expression of isoforms 4 and 5 were decreased in NRAS-mutant tumors, although this trend did not achieve statistical significance, as the proportion of these isoforms expressed at high levels was also decreased in NRAS-wt tumors. We plan to evaluate whether the other 3 isoforms that do not contain codon 61 cooperate with mutated isoform 1 or 2 to further augment or perhaps conversely neutralize the activation induced by the mutation. It will also be important to further characterize these isoforms in a non-BRAF mutated melanoma model, as the A375 cell line utilized in this study contains a BRAFV600E mutation.
As RAS has been deemed “undruggable” by many, the discovery of novel splice variants such as these NRAS isoforms may provide a new opportunity for therapeutic strategies [22, 23]. Isoform 2, in particular, with the additional exon 3b, could be an interesting target as isoform 2 appears to enhance melanoma proliferation and invasion, as well as resistance to vemurafenib treatment. Isoform 5 also appeared to enhance vemurafenib resistance, although the mechanism for this resistance, and thus how to overcome it, will require further investigation. Additionally, as current melanoma targeted therapies are now focused on inhibiting the downstream signal transduction pathways activated by oncogenes such as NRAS, a thorough understanding of which signaling pathways are activated by each of the isoforms may assist in the correct selection of targeted therapeutics when the levels of each isoform in a specific tumor are known.
In summary, this study demonstrates that the recently identified NRAS splice isoforms are expressed and biologically active in melanoma, and that their functions are distinct from one another. Additional investigation is needed to better characterize the downstream effects of NRAS isoform signaling, as the isoforms appear to vary significantly in their activation of downstream pathways. Importantly, the resistance to vemurafenib treatment seen with over-expression of isoforms 2 or 5 warrants potential consideration of these isoforms as biomarkers for therapeutic response, and the correlation of isoform 5 levels in metastases with enhanced survival indicates this isoform may have prognostic value in melanoma. Thus, further investigations into the role of these isoforms in melanoma will greatly benefit the biological and clinical understandings of this disease.
Supplementary Material
Grant Support:
This work was supported by NIH Grants: P01CA095426 (WEC), P30CA016058 (WEC), T32GM068412 (MCD).
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Mayo Clinic Staff (2016) Melanoma - Mayo Clinic. http://www.mayoclinic.org/diseases-conditions/melanoma/basics/definition/con-20026009. Accessed 18 Feb 2017
- 2.The American Cancer Society (2017) Key Statistics for Melanoma Skin Cancer. https://www.cancer.org/cancer/melanoma-skin-cancer/about/key-statistics.html. Accessed 18 Feb 2017
- 3.American Cancer Society (2016) Cancer Facts and Figures 2016. http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf. Accessed 26 May 2016
- 4.Lee C-S, Thomas CM, Ng KE (2017) An Overview of the Changing Landscape of Treatment for Advanced Melanoma. Pharmacother J Hum Pharmacol Drug Ther. doi: 10.1002/phar.1895 [DOI] [PubMed] [Google Scholar]
- 5.McCormick F (1995) Ras-related proteins in signal transduction and growth control. Mol Reprod Dev 42:500–506. doi: 10.1002/mrd.1080420419 [DOI] [PubMed] [Google Scholar]
- 6.Curtin JA, Fridlyand J, Kageshita T, et al. (2005) Distinct sets of genetic alterations in melanoma. N Engl J Med 353:2135–47. doi: 10.1056/NEJMoa050092 [DOI] [PubMed] [Google Scholar]
- 7.Albino AP, Le Strange R, Oliff AI, et al. Transforming ras genes from human melanoma: a manifestation of tumour heterogeneity? Nature 308:69–72. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinopoulos PA, Karamouzis MV., Papavassiliou AG (2007) Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov 6:541–555. doi: 10.1038/nrd2221 [DOI] [PubMed] [Google Scholar]
- 9.Fedorenko IV, Gibney GT, Smalley KSM (2013) NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene 32:3009–18. doi: 10.1038/onc.2012.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–55. doi: 10.1038/nrg2776 [DOI] [PubMed] [Google Scholar]
- 11.Wang ET, Sandberg R, Luo S, et al. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476. doi: 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djebali S, Davis CA, Merkel A, et al. (2012) Landscape of transcription in human cells. Nature 489:101–108. doi: 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Coulombe-Huntington J, Kang S, et al. (2016) Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 164:805–817. doi: 10.1016/j.cell.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisfeld A-K, Schwind S, Hoag KW, et al. (2014) NRAS isoforms differentially affect downstream pathways, cell growth, and cell transformation. Proc Natl Acad Sci U S A 111:4179–84. doi: 10.1073/pnas.1401727111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markowitz J, Mal TK, Yuan C, et al. (2016) Structural characterization of NRAS isoform 5. Protein Sci 25:1069–1074. doi: 10.1002/pro.2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–4. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts A, Pachter L (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10:71–3. doi: 10.1038/nmeth.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin del Campo SE, Latchana N, Levine KM, et al. (2015) MiR-21 Enhances Melanoma Invasiveness via Inhibition of Tissue Inhibitor of Metalloproteinases 3 Expression: In Vivo Effects of MiR-21 Inhibitor. PLoS One 10:e0115919. doi: 10.1371/journal.pone.0115919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burd CE, Liu W, Huynh MV., et al. (2014) Mutation-Specific RAS Oncogenicity Explains NRAS Codon 61 Selection in Melanoma. Cancer Discov 4:1418–1429. doi: 10.1158/2159-8290.CD-14-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandalà M, Merelli B, Massi D (2014) Nras in melanoma: Targeting the undruggable target. Crit Rev Oncol Hematol 92:107–122. doi: 10.1016/j.critrevonc.2014.05.005 [DOI] [PubMed] [Google Scholar]
- 23.Posch C, Ortiz-Urda S (2013) NRAS mutant melanoma – undrugable? Oncotarget 4:494–495. doi: 10.18632/oncotarget.970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


