Abstract
During protein synthesis, the mRNA helicase activity of the ribosome ensures that codons are made single-stranded before decoding. Here, based on recent structural and functional findings, a quantitative model is presented for a tandem arrangement of two helicase active sites on the ribosome. A distal site encounters mRNA structures first, one elongation cycle earlier than a proximal site. Whereas unwinding of encountered mRNA structures past the proximal site is required for translocation, two routes exist for translocation past the distal site: sliding, which requires unwinding, and stick-slip, which does not. The model accounts in detail for a number of findings related to the ribosomal helicase and provides a testable framework to further study mRNA unwinding.
INTRODUCTION
During translation, the ribosome translocates along a messenger RNA (mRNA) in the 5’-to-3’ direction in steps of three nucleotides (nt), and recruits transfer RNAs (tRNAs) carrying amino acids at each step. A translocating bacterial ribosome unwinds encountered folded mRNA segments, and is therefore also an mRNA helicase [1–3] despite no homology to previously known nucleic acid helicase superfamilies [4]. Its helicase active site has been localized to the vicinity of the entrance to the downstream mRNA tunnel between the head and body domains of the small (30S) ribosomal subunit [1,2,5]. The entrance is lined by ribosomal protein S3 from the head domain and ribosomal proteins S4 and S5 from the body domain (Fig. 1A), and can accommodate only a single strand of mRNA. Mutation of S3 and S4 residues surrounding the tunnel entrance diminish the activity of the helicase [2], implicating these proteins in its mechanism of action.
Fig. 1. Binding of single-stranded mRNA to ribosomal protein S3 outside of the mRNA tunnel entrance [9].
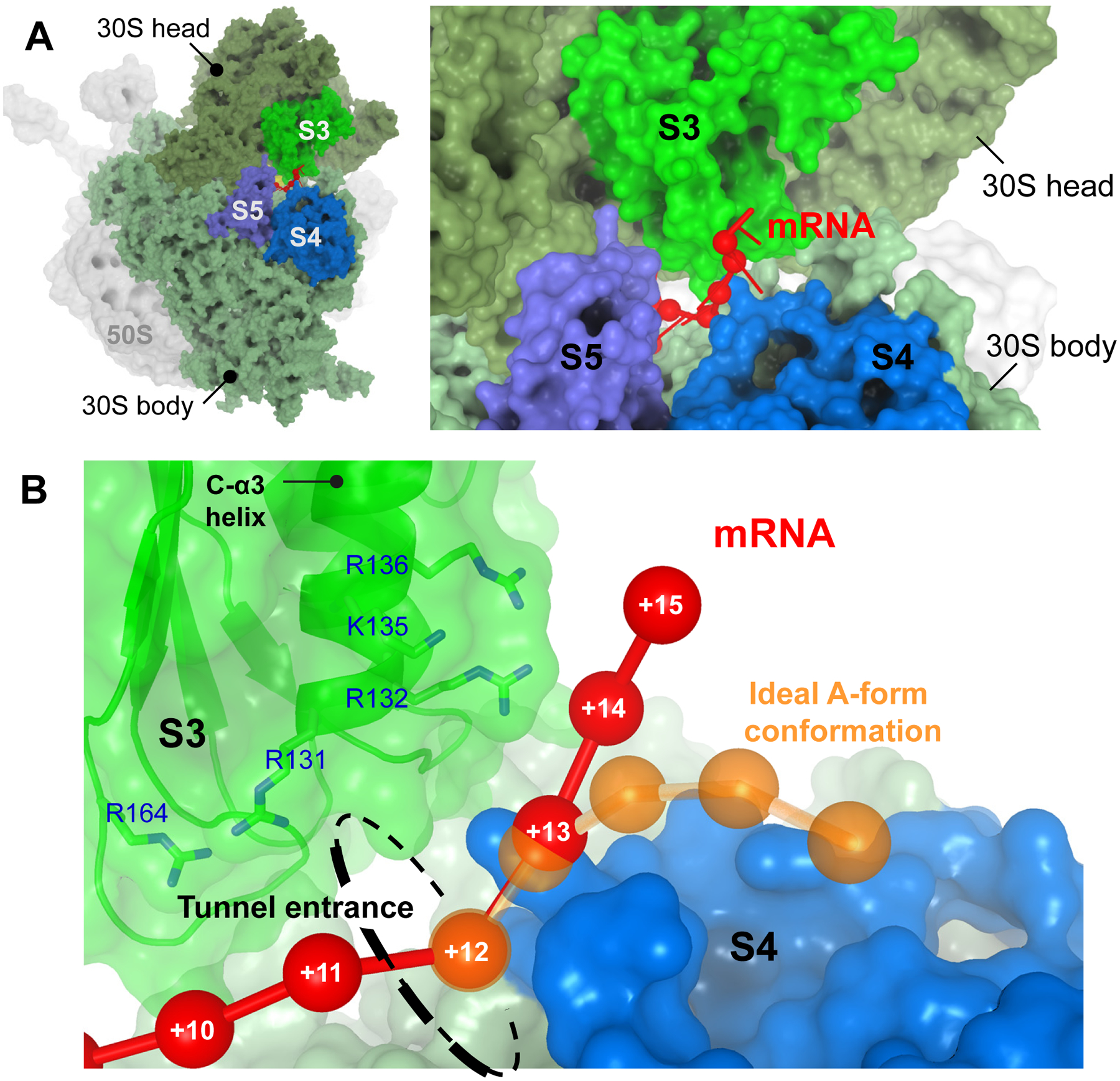
A. The 3’ end of the mRNA (red) emerges at the tunnel entrance between the head and the body domain of the small ribosomal subunit, and interacts mainly with ribosomal protein S3 outside of the tunnel. B. The path of the S3-bound single-stranded mRNA (red) deviates from that of an ideal A-form strand (orange). The phosphorus atoms of mRNA nucleotides are represented by spheres and are numbered starting from the first nucleotide of the P-site codon.
Translocation is catalyzed by elongation factor G (EF-G) and involves rotation of the 30S head domain relative to the body domain by ~20° around the neck axis [6]. First, “forward” head rotation moves tRNAs and the mRNA relative to the body domain, resulting in the so-called chimeric hybrid state [7,8]. Next, “reverse” head rotation must occur, during which the mRNA and tRNAs would no longer follow the movement of the head, but rather hold on to the body as the head moves back. Reverse head rotation results in the classical-state post-translocation ribosome.
Structural data indicate that the mRNA tunnel is lengthened upon forward head rotation, enclosing an additional 3 nt of downstream mRNA [9]. For a tunnel that can accommodate only a single strand, such a lengthening will occur only if the 3-nt segment is unwound. This describes a “passive” helicase mechanism whereby translocation is delayed by spontaneous unwinding of encountered mRNA structures. In contrast, an “active” helicase would destabilize the folding of such structures before translocating over them, and so its translocation rate would be less compromised [10–12]. Single-molecule kinetic measurements using optical tweezers have made it clear that the ribosomal helicase is highly active, appearing to use not one but two distinct active helicase mechanisms [1]. It is not known what these mechanisms are and what their structural basis is.
How can the highly active character of the ribosomal helicase be explained? How does the structure of the tunnel entrance support such an active character, and what role(s) do the proteins surrounding the entrance play? How do forward and reverse 30S head rotations during translocation contribute to unwinding? Finally, what evolutionary adaptations have allowed the ribosome to unwind mRNAs so efficiently yet frugally? Here, we present a model of tandemly-arranged helicase sites on the ribosome that addresses these important questions. It is introduced qualitatively based on structural observations, and quantitatively in the form of a kinetic scheme. The model accounts for a number of experimental observations and makes further testable predictions.
MODEL OUTLINE
Recent structural studies have revealed the binding of 3 nucleotides of single-stranded mRNA to ribosomal protein S3 just outside of the tunnel [9,13]. The backbone of the bound mRNA is extended and straightened, deviating significantly from the curved A-form geometry (Fig. 1B). The deviation indicates a binding specificity for the single-stranded form of mRNA, since double-stranded RNA cannot adopt the observed configuration (Supp. Note 1). The bound single-stranded mRNA segment can be considered a helicase product stabilized by the binding interaction, similar to product stabilization by DEAD-box RNA helicases [14]. In other words, the helicase acts to disfavor structured mRNA segments, by stabilizing their single-stranded state, before the structures reach the entrance itself.
This leads to the present proposal of two tandem “active sites” where mRNA unwinding can occur (Fig. 2A). mRNA structure is first encountered at a distal active site, and if not unwound, arrives in the next elongation cycle at a proximal active site (Fig. 2B,C). If we define the first nucleotide of the P-site codon inside the ribosome as position +1, crystal structures place the proximal and distal active sites near positions +11 and +14, respectively (Fig. 2A). The proximal active site corresponds to the tunnel entrance in the classical state [15], matching the position of the helicase active site predicted from biochemical analysis [2]. The distal active site is immediately downstream from the S3-bound single-stranded mRNA [9], near the 3’ end of the ribosomal footprint on mRNA [16,17]. We refer to the corresponding mRNA segments acted upon by the proximal and distal active sites as the proximal segment (positions +12 to +14) and the distal segment (+15 to +17), respectively (Fig. 2A).
Fig. 2. The tandem active site model for the ribosomal helicase.
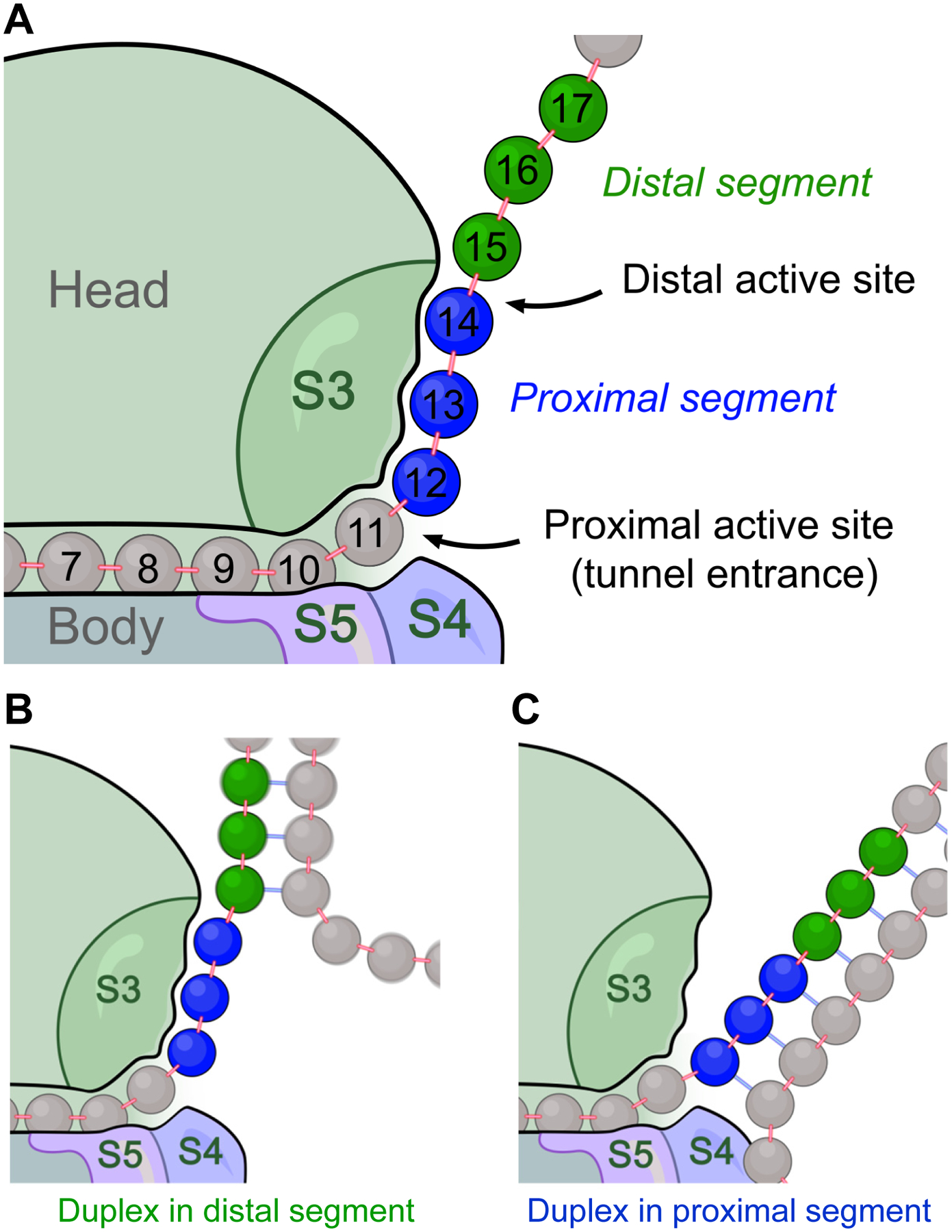
A. Cartoon showing the tandem arrangement of helicase active sites on the ribosome. The proximal and distal active site correspond to positions +11 and +14, respectively. The proximal segment of mRNA (positions +12–14; blue) is shown in its S3-bound state, and the distal segment (positions +15–17; green) is shown in its open state. B and C. Encounter between a downstream mRNA duplex and the distal active site (in B) or the proximal active site (in C). During translation, encounter by the distal active site occurs first. Encounter by the proximal active site occurs in the next elongation cycle, after the distal active site fails to unwind the duplex.
Unwinding of the proximal segment is required for translocation to proceed, while unwinding of the distal segment is optional (Fig. 3). More specifically, forward head rotation requires unwinding of the proximal segment (Fig. 3A) due to the tunnel lengthening mentioned earlier; reverse head rotation can then occur by two alternative routes, only one of which requires unwinding of the distal segment (Fig. 3B).
Fig. 3. Fate of translocation upon encountering a duplex.
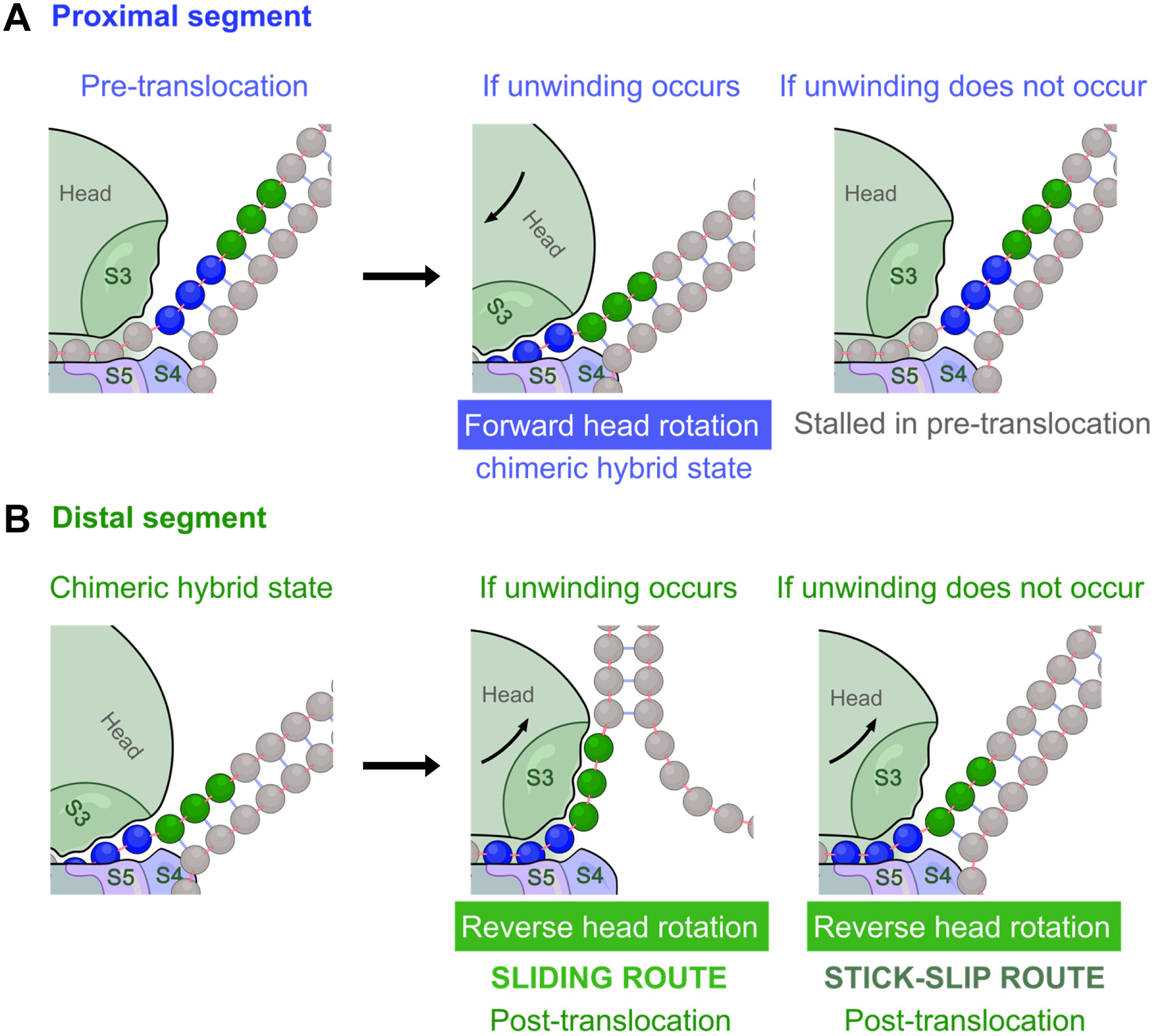
A and B. Cartoons showing encounters between downstream helical mRNA elements and the proximal active site (in A) or distal active site (in B). For each active site, the starting state (left panel), and the outcome with and without unwinding (middle and right panels, respectively) are shown. A. At the proximal active site, translocation (by forward head rotation) can proceed only if the proximal segment (blue) unwinds spontaneously. If unwinding does not occur, the ribosome will remain stalled in the pre-translocation state. B. At the distal active site, if unwinding of the distal segment (green) occurs, S3 can slide over the unwound segment during reverse head rotation. If no unwinding occurs, the distal segment can still “slip” through this active site during reverse head rotation, resulting in loss of S3-mRNA binding. Either of these scenarios (sliding or stick-slip) at the distal active site successfully translocates mRNA by 3 nucleotides and arrives at the post-translocation state.
During reverse head rotation, the 30S head (which contains protein S3) must move relative to mRNA. This movement can be accomplished by either sliding or stick-slip at the distal active site (Fig. 3B). Sliding refers to relative movement of S3 from the proximal onto the distal segment without dissociating from the mRNA, an isoenergetic diffusion process that can be very fast [18]. It can occur only if the distal segment is single-stranded. In contrast, stick-slip involves the uncompensated dissociation of the proximal mRNA segment from S3. It can occur whether or not the distal segment is single-stranded, meaning that stable mRNA structures can take this route to skip the distal active site and reach the proximal site. A simplified animation for this model is shown in Supp. Movie 1.
A kinetic scheme based on this model is described below. It accounts for the force-dependent rates of ribosomal translocation over mRNA duplexes observed in single-molecule optical tweezers experiments [1].
KINETIC SCHEME
In each elongation cycle, consider the proximal segment of mRNA (+12 to +14) to be in one of three states: closed, open, or bound (Fig. 4A), whereas the distal segment (+15 to +17), can be either closed or open (Fig. 4B). The Boltzmann factors for the partition functions describing the probability of these states are listed in Table 1.
Fig. 4. The main states of (A) the proximal segment, and (B) the distal segment of the mRNA outside of the tunnel entrance.
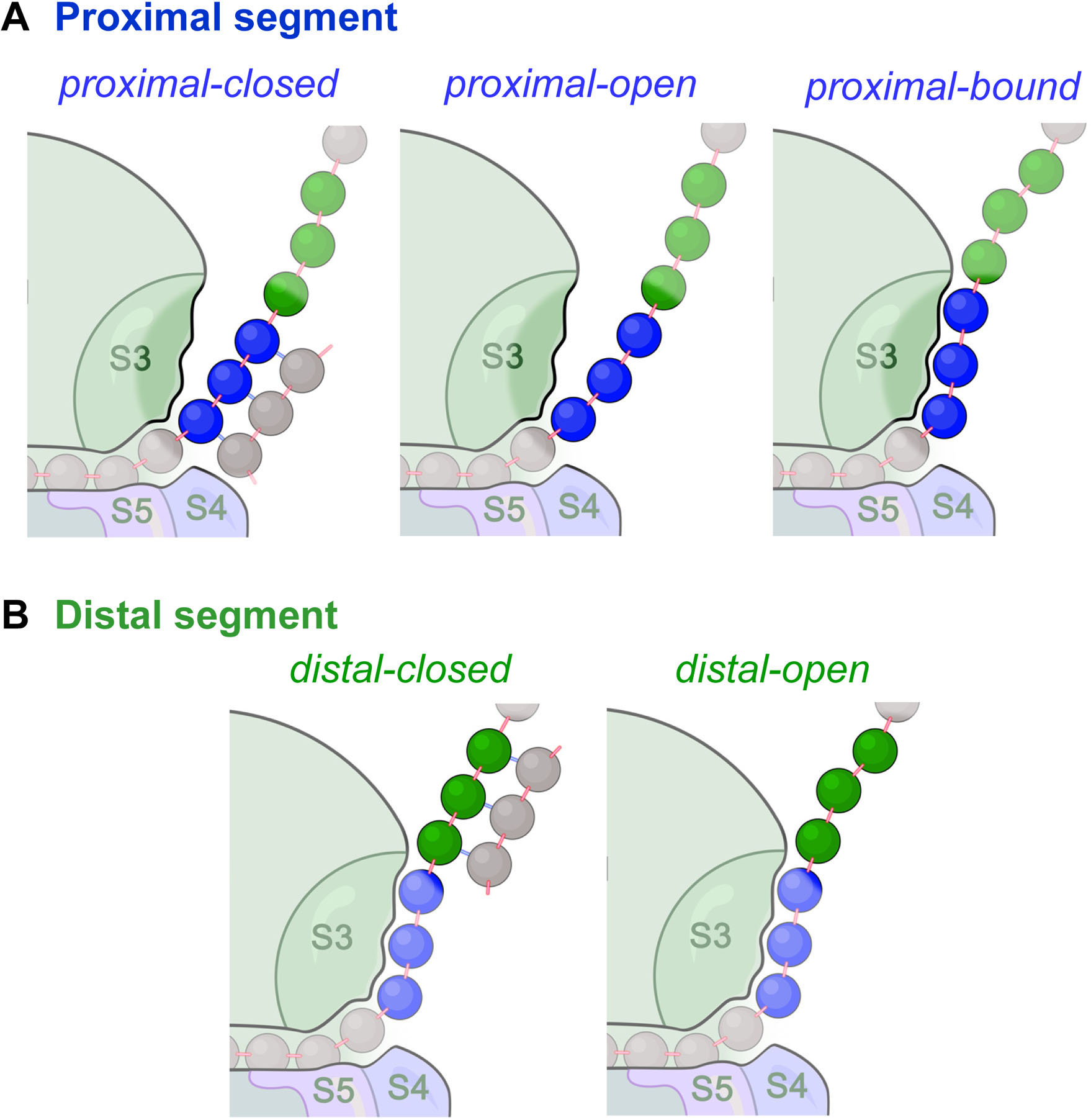
A. The proximal segment (blue) can be in one of three states (closed, open, or bound). B. The distal segment (green) can be in one of two states (closed or open).
Table 1.
Boltzmann factors for the proximal and distal segments of mRNA in the Gibbs ensemble
| Segment and state | Boltzmann factor a |
|---|---|
| Proximal-open | exp(m[FXF – ΔGstretch] / kBT) |
| Proximal-closed | exp(mΔGbp / kBT) |
| Proximal-bound | exp(mΔGbinding / kBT) |
| Distal-open | exp(s[FXF – ΔGstretch] / kBT) |
| Distal-closed | exp(sΔGbp / kBT) |
exp denotes the exponential function. m=3 is the length of the proximal segment of mRNA [9], and s = 3 is the translocation step size for the ribosome dictated by codon size. F is the magnitude of applied external pulling force on the mRNA ends (e.g. by optical tweezers), and XF is the extension of single-stranded RNA at each applied force F according to the worm-like chain (WLC) model [33–35] (Supp. Note 3). The free energies are chosen to be positive: ΔGstretch is the stretching work for single-stranded mRNA per opened base pair in the WLC model [33], ΔGbp is the average base pair stability at zero force according to nearest neighbor rules [36], and ΔGbinding (the only free parameter in this table) is the average mRNA-S3 binding stability per nucleotide at zero force. kB is the Boltzmann constant, and T is temperature.
The kinetic scheme is shown in Fig. 5A for one elongation cycle. The overall unidirectional translation rate is:
| (1) |
where ktranslocation is the effective rate of the translocation step, and kdwell is the apparent rate combining all other events of an elongation cycle (tRNA accommodation, peptide bond formation, etc.)
Fig. 5. The kinetic scheme for the tandem active site model.
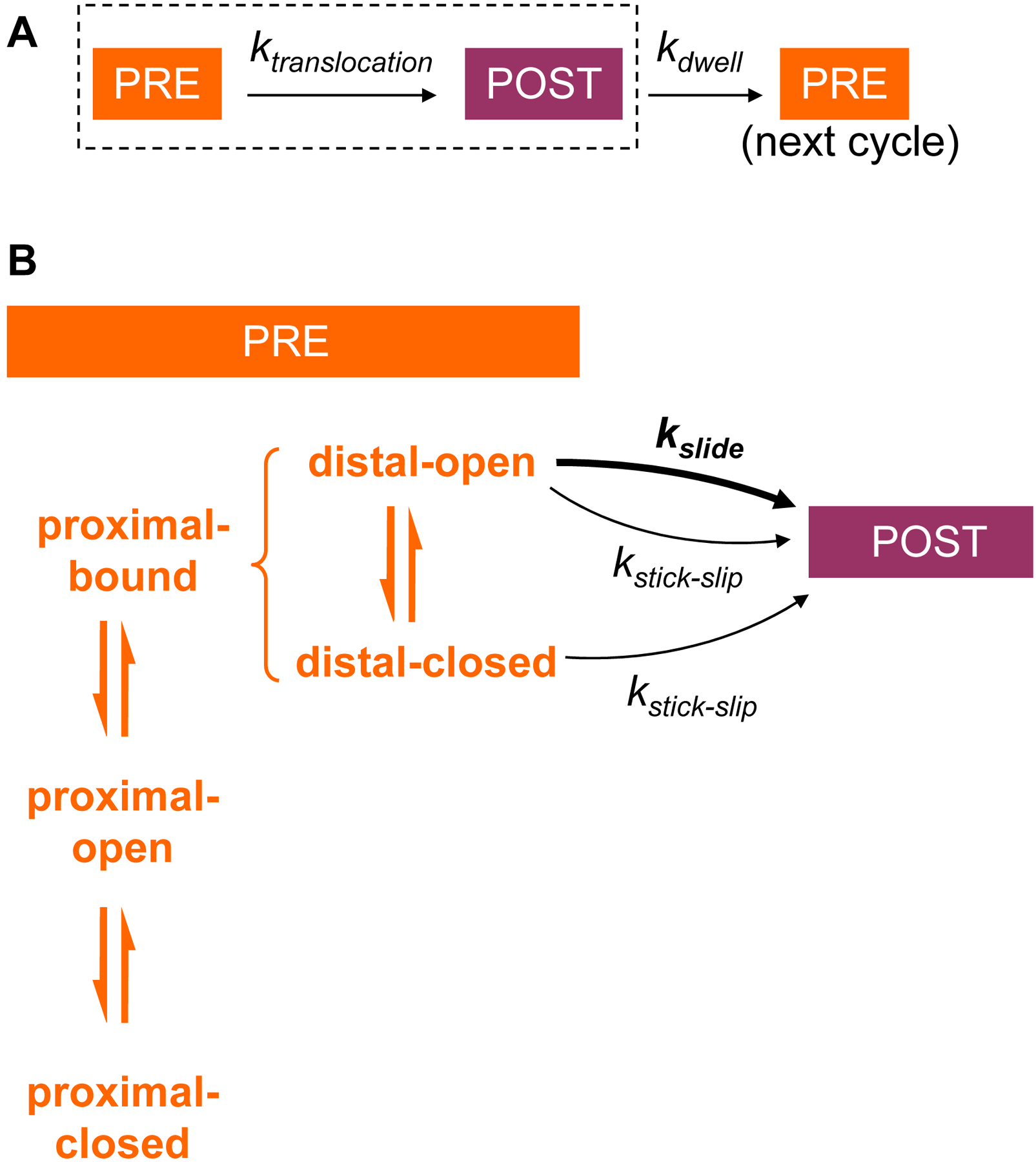
A. The translation elongation pathway is composed of translocation and dwell steps. PRE and POST denote pre-translocation and post-translocation states of the ribosome complex, respectively. B. The translocation step of panel A is expanded to show the pre-translocation equilibrium in the proximal and distal segments, indicated on the left. The rate of translocation depends on mRNA duplex stability in the proximal segment (affecting the probability of the proximal-bound state), and on mRNA duplex stability in the distal segment (affecting probability of the distal-open state). To be able to translocate at all, the system has to be in the proximal-bound state. To be able to proceed by rapid sliding, it further has to be in the distal-open state. Stick-slip is slow, but can occur from either state of the distal segment. After translocation and during the time controlled by kdwell, the new proximal segment can relax to equilibrium between its three states prior to the next translocation step.
The translocation rate can be written as:
| (2) |
where Pproximal-bound is the probability of the bound state of the proximal segment, and ktranslocation-bound is the rate of translocation starting from this state (see Supp. Note 2).
From this state (Fig. 6A), forward head rotation is unhindered and simply moves both S3 and its bound mRNA together (Fig. 6B). Next, in reverse head rotation, S3 and mRNA move relative to each other via either sliding or stick-slip (Fig. 6C,D). Since sliding is possible only for the open state of the distal segment, its effective rate is limited by Pdistal-open, the probability of this state. The net rate of the two routes is therefore the sum:
| (3) |
where kslide and kstick-slip are the intrinsic rates of the sliding and stick-slip routes, respectively. (Pdistal-open kslide is the effective rate of the sliding route.)
Fig. 6. Translocation over an mRNA hairpin encountered at the distal active site.
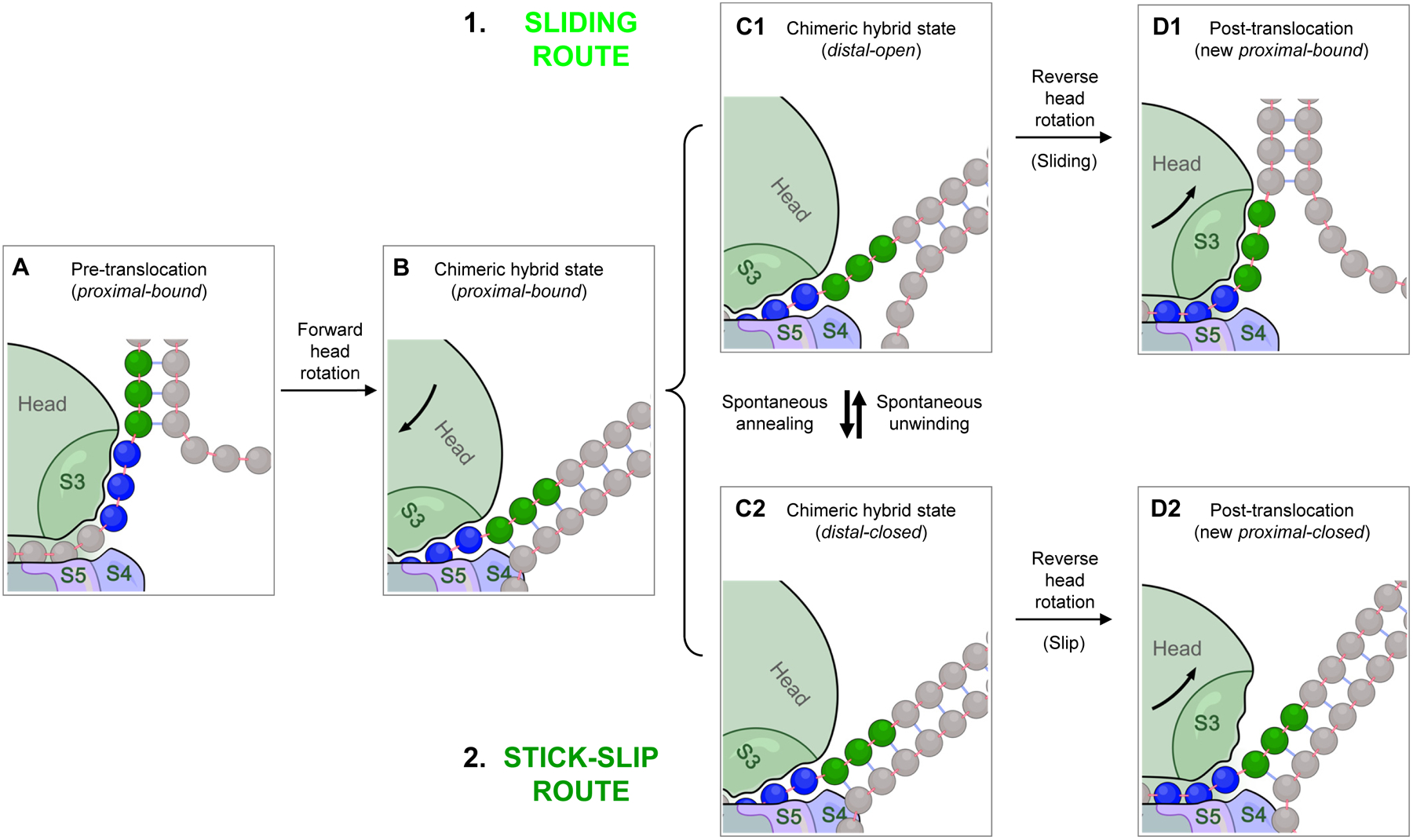
A. Before translocation starts, the proximal mRNA segment (blue) must be in the bound state. B. Forward head rotation moves S3 (part of the head domain) along with its bound mRNA as a rigid body relative to the body domain, with no unwinding. C1 and C2. The head-rotated intermediate in panel B is in fact composed of two different states of the distal segment (green) in equilibrium: (C1) the distal-open state, and (C2) the distal-closed state. The same equilibrium exists in the pre-translocation state (panel A), but not shown for simplicity. D1 and D2. During reverse head rotation, the head domain moves back to its classical state, whereas the mRNA remains in place, forcing S3 to move relative to the mRNA. If the distal segment is open, S3 can slide along the mRNA, resulting in a post-translocation state in which the now-proximal segment is in the bound state (D1). If the distal segment is closed, S3 loses its bound mRNA without acquiring compensatory binding; i.e. stick-slip occurs, resulting in a proximal-closed post-translocation state (D2). The latter has to spontaneously convert to proximal-bound state before the next translocation step can take place. Note that the distal-open state (panel C1) can also undergo stick-slip to yield a proximal-open post-translocation product (not shown; see Fig. 5), but this occurs relatively rarely for this state, and omitted here for simplicity.
Sliding maintains mRNA-S3 contact whereas stick-slip does not. The rate constants kslide and kstick-slip can therefore be related by the stability of S3-mRNA binding for each new encountered nucleotide (ΔGbinding):
| (4) |
Using the published rates of translation with Escherichia coli ribosomes measured under different applied optical tweezers pulling forces at room temperature [1], the values of the three free parameters used in the kinetic scheme were estimated (Table 2). With these values, the translation rates as a function of the applied force calculated from Eq. 1 are in excellent agreement with the experimental measurements made on mRNA hairpins of either 50% or 100% GC content (Fig. 7), which differ only in their base-pair stabilities (ΔGbp).
Table 2.
Parameters of the kinetic scheme obtained by fitting to optical tweezers data [1]
| Parameter | Description | Value |
|---|---|---|
| ΔGbinding | mRNA-S3 binding stability | 3 kcal/mol (per nucleotide) |
| kstick-slip | Rate of the stick-slip route | 0.47 s−1 (codons per second) |
| kdwell | Rate of non-translocation events | 0.43 s−1 (codons per second) |
Fig. 7. Agreement between the model and experimental measurements.
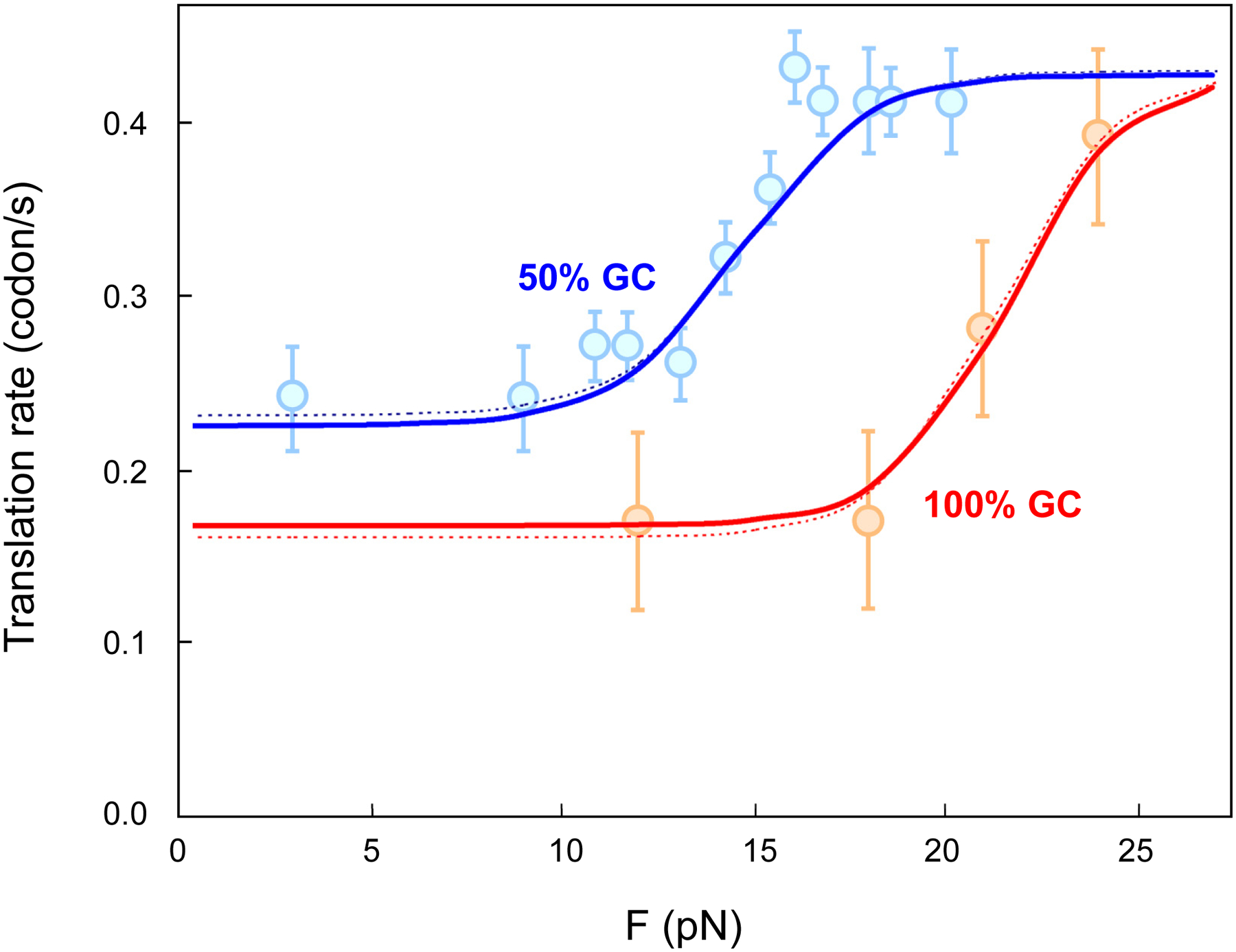
Translation rates predicted by the kinetic scheme for the tandem active site model, and the measured translation rates under different optical tweezers pulling forces [1] are shown for unwinding of hairpins with 50% and 100% GC content (blue and red curves, respectively). Each data point shows the mean measured rate and the standard error of the mean. The dotted curves are reproduced from the fitting done in the original study [1]. The complete set of parameter values and formulas are listed in Supp. Note 3. Data points adapted from [1] with permission.
The effect of varying base-pair stability on predicted translation rates according to this scheme is plotted in Fig. 8. For reverse head rotation, the fast sliding route prevails when encountering weakly base-paired structures, while the slow stick-slip route becomes dominant with more stable structures. At yet higher barrier stabilities, forward head rotation is also impeded and can eventually become rate-limiting.
Fig. 8. Dependence of translation rates on base-pair stability (ΔGbp) of encountered mRNA segments.
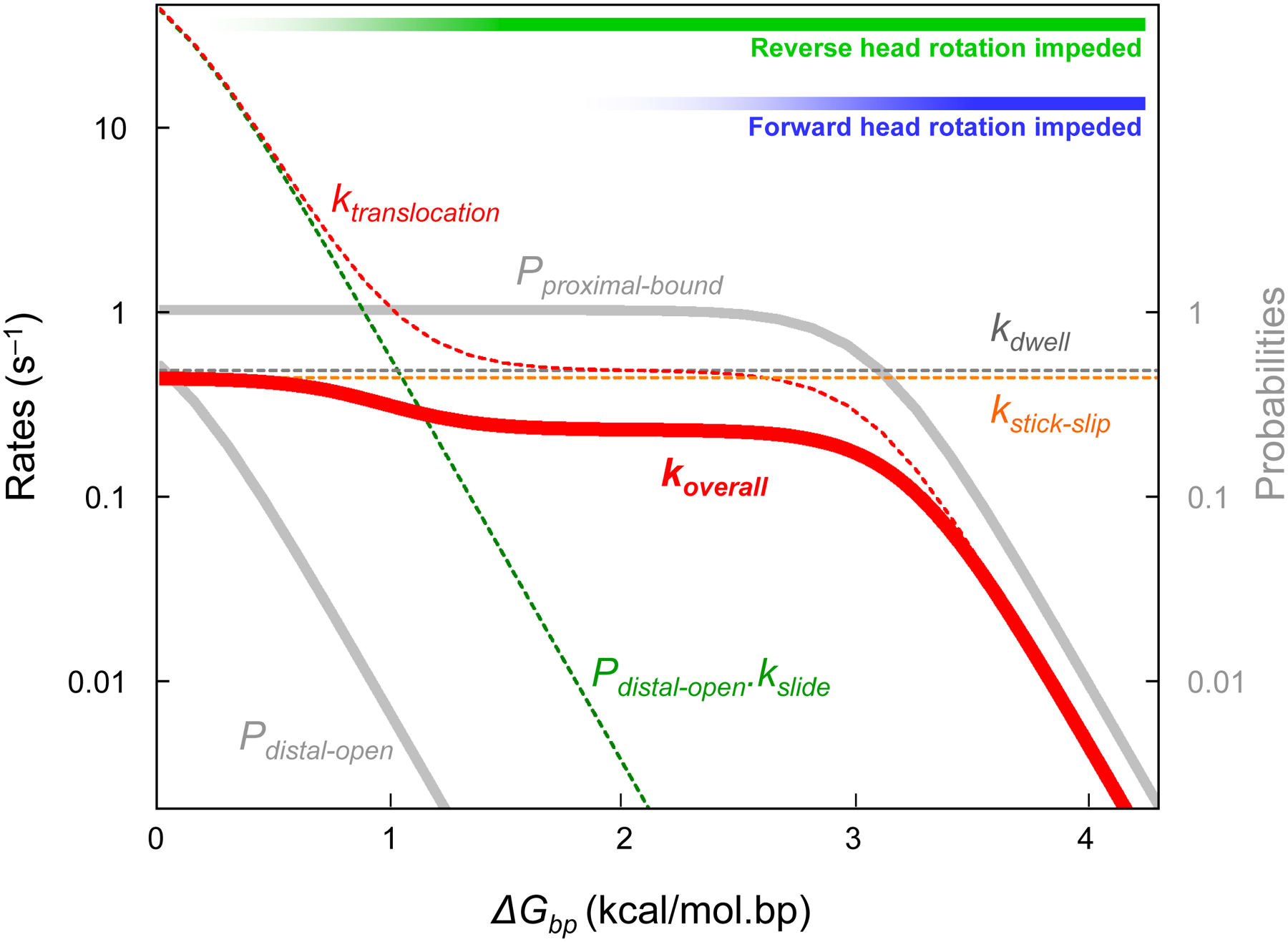
Various rates and probabilities in the kinetic scheme are plotted in logarithmic scale as a function of ΔGbp. Three kinetic regimes can be distinguished: at low values of ΔGbp, translocation is not rate-limiting, and reverse head rotation occurs mostly by sliding; At intermediate ΔGbp, reverse head rotation becomes rate-limiting as the slow stick-slip route predominates. At yet higher ΔGbp, even forward head rotation becomes hindered.
The formulas used in the kinetic scheme are summarized in Supp. Note 3. The sensitivity of the scheme to the choice of Boltzmann factors is discussed in Supp. Note 4.
DISCUSSION
Based on clues from structural observations (Fig. 1) [9,13], the proposed model for the ribosomal helicase envisions two active sites operating in tandem to unwind encountered mRNA structures (Figs. 2,3) and forms the basis for a kinetic scheme for translocation over these structures (Fig. 5). The kinetic scheme can account for the rates of ribosome translocation through mRNA hairpins as measured by optical tweezers (Fig. 7), using only three free parameters (Table 2).
The S3-mRNA binding energy (~9 kcal/mol per codon) is a key parameter in the kinetic scheme, controlling not only the destabilization of encountered structures at the proximal active site (Table 1), but also the relative rates of the sliding and stick-slip routes at the distal active site (Eq. 4). Importantly, to dissociate mRNA from S3 in the stick-slip route at a significant rate, the ribosome must have evolved to perform work that exceeds (or is within a few kBT of) this binding energy. The main energy sources for translocation are peptidyl transfer and GTP hydrolysis by EF-G [19], the latter alone being ~12 kcal/mol [20], suggesting that stick-slip is indeed affordable by the ribosome under this model.
The model predicts that placing a hairpin at the tunnel entrance would tend to favor melting of its first three base pairs. The now-unpaired nucleotides of the 5’ strand of the stem would become immobilized by binding to protein S3, while those of the 3’-strand of the stem would become flexible. Consistent with this prediction, chemical probing of an mRNA hairpin placed at the tunnel entrance revealed low backbone flexibility in the first 3–4 nt of the 5’ strand, but high backbone flexibility in the complementary 3’-strand nucleotides [13].
According to this model, a stable structure can hinder translocation in two ways: in the distal segment, it can delay reverse head rotation until either unwinding or stick-slip occurs; in the proximal segment, if sufficiently stable, it can delay forward head rotation until unwinding occurs (Fig. 8, top right). Consequently, while a moderately stable structure in the distal segment would likely follow the sliding route after unwinding at the distal active site, a highly stable structure would likely take the slow stick-slip route at this site and unwind mainly at the proximal active site in the next elongation cycle. Furthermore, since E-site tRNA release correlates with completion of reverse head rotation [6,21], a stable structure can slow down tRNA release when in the distal segment. These predictions are in striking agreement with results of a study in which fluorescence and single-molecule Förster resonance energy transfer (smFRET) between fluorophores placed on different parts of the ribosome complex were used to monitor translocation on mRNAs with hairpins of either 65% or 100% GC content starting at position +15 [22]. The 100% GC hairpin first slowed down E-site tRNA release and then tRNA translocation, while the 65% GC hairpin at most only slowed down E-site tRNA release (Fig. 9). Delay in E-site tRNA release due to a pseudoknot barrier has also been detected by bulk kinetic analysis [23]. These and other studies on frameshift-inducing mRNA structures point to a possibly complex non-canonical chain of events during −1 frameshifting that change 30S-50S inter-subunit rotational dynamics as well as 30S head dynamics [24–27]. Frameshifting may proceed in degenerate pathways [28] with multiple potential pausing steps as the ribosome approaches a barrier [23,24], a notion consistent with the present model.
Fig. 9. Explaining the effect of mRNA secondary structures on translation rates using the model.
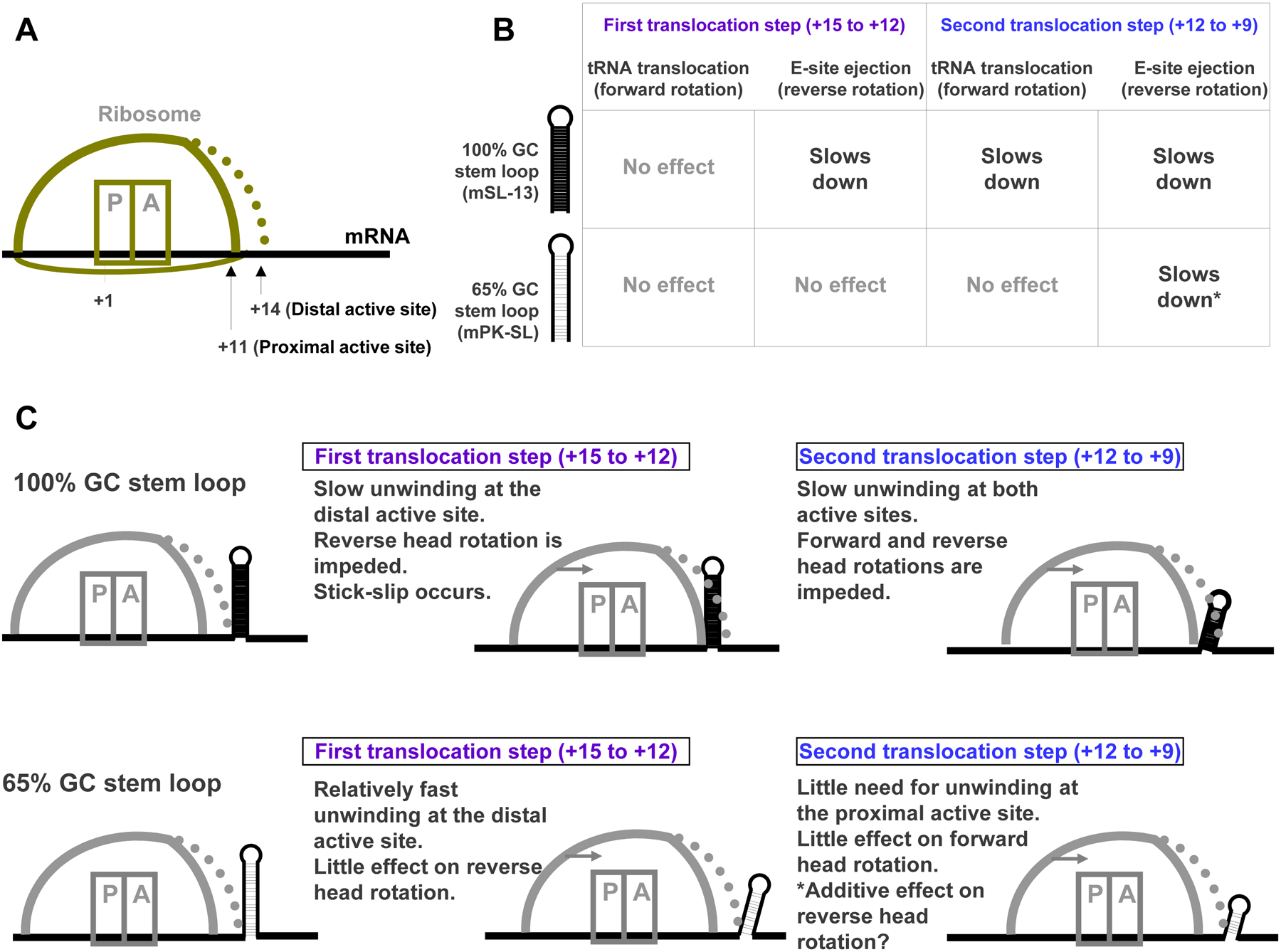
A. Cartoon representing the ribosome and its proximal and distal helicase active sites. B. Summary of experimental data [22] on the effect of two stem-loop structures, with 100% and 65% GC content, on translocation rates (adapted from Table S2 in the reference). C. Predictions of the tandem active site model for the rates of the steps outlined in panel B.
In typical optical tweezers experiments, translation of every additional codon in a structured mRNA by the ribosome requires the opening of 3 base pairs of a hairpin, releasing two 3-nt single strands and increasing the end-to-end extension of mRNA by a total of 6 nt [3]. Interestingly, the sliding and stick-slip routes in the present model are predicted to result in different changes in the end-to-end extension. Whereas sliding would produce a single 6-nt step corresponding to the opening of the distal segment and its complement (Fig. 10A), stick-slip would produce two ~3-nt steps, one from the slip event itself (with no unwinding), and one from the following equilibrium unwinding and binding of the segment (Fig. 10B) (Supp. Note 5). Such 3-nt ribosomal “substeps” have indeed been detected recently using high-resolution optical tweezers instrumentation [29]. The substeps are predicted to occur more frequently on more stable hairpins.
Fig. 10. Prediction of end-to-end mRNA extension in optical tweezers experiments based on the model.
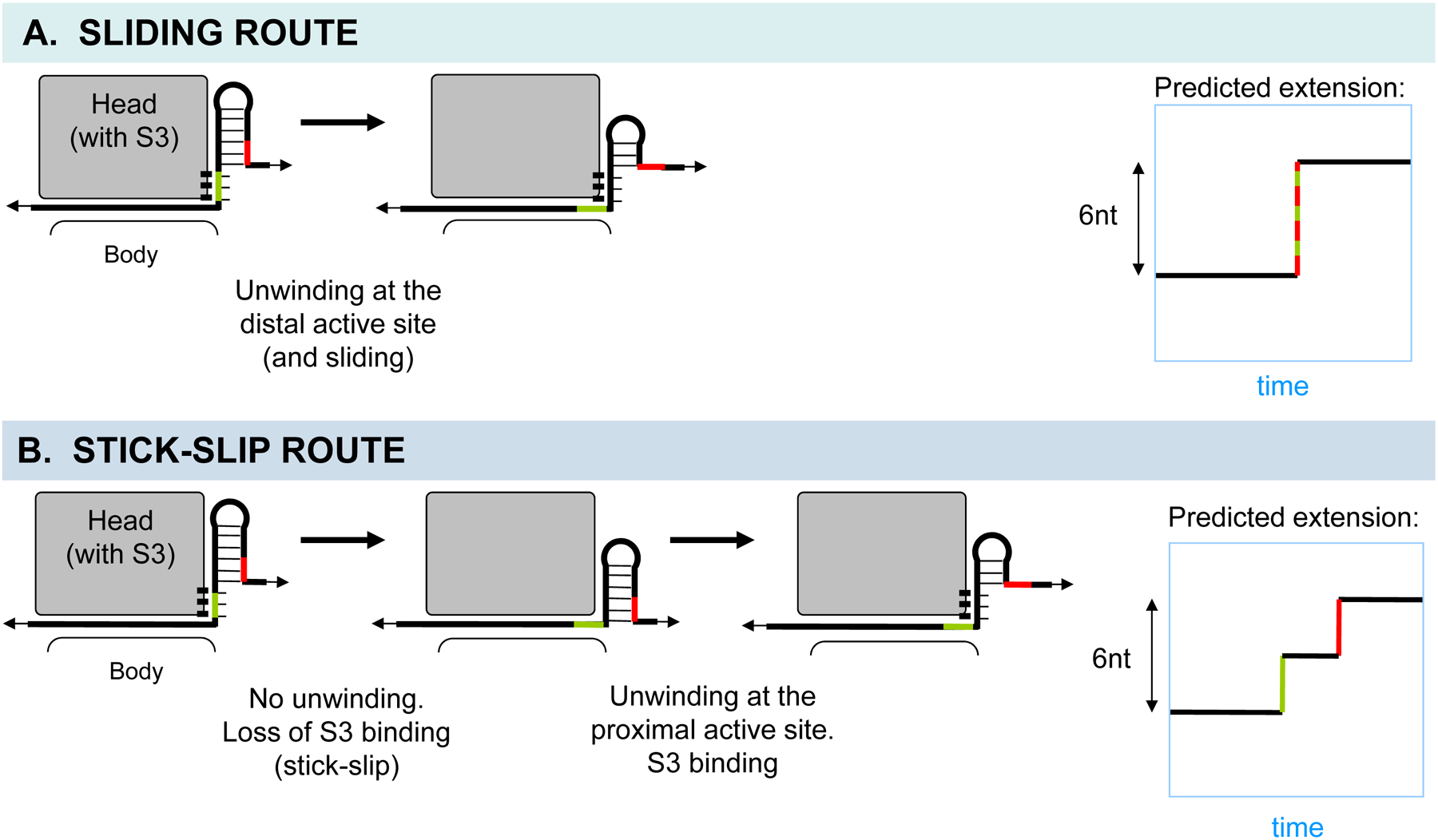
A and B. The head domain of the small subunit is shown in gray, and the binding between the proximal segment of mRNA and S3 is indicated. A. The sliding route generates a major 6-nt step due to unwinding of the distal segment during translocation. B. The stick-slip route can generate two distinct ~3-nt steps, one due to slip itself during translocation, and the other due to unwinding and equilibrium binding of the new proximal segment after translocation. The ratio of the two substep sizes depends on the angle of the S3-mRNA interface with respect to the pulling force. Also see Supp. Note 5.
A somewhat unexpected prediction of the tandem active site model is that reverse head rotation would be hindered when the 3’ terminus of an mRNA is encountered at the distal active site (or further inside the ribosome). This is because the lack of a distal segment eliminates the sliding route, and a slow stick-slip-like event has to occur to allow mRNA movement relative to the head. This prediction has not been addressed directly. In fact, careful kinetic measurements of reverse head rotation have been performed using short mRNAs whose 3’ termini are at position +14 or closer, yielding an estimated rate of ~10–15 s–1 [21,30–32]. It is possible that these measurements reflect reverse head rotation hindered in part by proximity of the 3’ terminus.
The tandem active site model and its kinetic scheme presented here are simplistic approximations of the mRNA unwinding process. In essence, the model describes a binding site for single-stranded mRNA on the ribosome, and a bifurcation into sliding and stick-slip regimes for mRNA movement over this site, a paradigm that may apply also to other active helicases.
Supplementary Material
Schematic animation of mRNA unwinding at the proximal and distal active sites.
ACKNOWLEDGMENTS
This work was supported by grant No. R35 GM118156 from the U.S. National Institutes of Health to H.F.N. We thank Clive Bagshaw for critical reading of the manuscript, and Carlos J. Bustamante for helpful discussions.
REFERENCES
- 1.Qu X, Wen J-D, Lancaster L, Noller HF, Bustamante C & Tinoco I (2011) The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takyar S, Hickerson RP & Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120, 49–58. [DOI] [PubMed] [Google Scholar]
- 3.Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, Noller HF, Bustamante C & Tinoco I (2008) Following translation by single ribosomes one codon at a time. Nature 452, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caruthers JM & McKay DB (2002) Helicase structure and mechanism. Current Opinion in Structural Biology 12, 123–133. [DOI] [PubMed] [Google Scholar]
- 5.Yusupova GZ, Yusupov MM, Cate JH & Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106, 233–241. [DOI] [PubMed] [Google Scholar]
- 6.Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN & Spahn CMT (2010) Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Lancaster L, Donohue JP & Noller HF (2013) Crystal Structures of EF-G-Ribosome Complexes Trapped in Intermediate States of Translocation. Science 340, 1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Lancaster L, Donohue JP & Noller HF (2014) How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amiri H & Noller HF (2019) Structural evidence for product stabilization by the ribosomal mRNA helicase. RNA 25, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betterton MD & Jülicher F (2005) Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys Rev E Stat Nonlin Soft Matter Phys 71, 011904. [DOI] [PubMed] [Google Scholar]
- 11.Lohman TM & Bjornson KP (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 12.Manosas M, Xi XG, Bensimon D & Croquette V (2010) Active and passive mechanisms of helicases. Nucleic Acids Res. 38, 5518–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Hong S, Ruangprasert A, Skiniotis G & Dunham CM (2018) Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel. Structure 26, 437–445.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder P & Jankowsky E (2011) From unwinding to clamping — the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12, 505–516. [DOI] [PubMed] [Google Scholar]
- 15.Jenner LB, Demeshkina N, Yusupova G & Yusupov M (2010) Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol 17, 555–560. [DOI] [PubMed] [Google Scholar]
- 16.Mohammad F, Woolstenhulme CJ, Green R & Buskirk AR (2016) Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep 14, 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steitz JA (1969) Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224, 957–964. [DOI] [PubMed] [Google Scholar]
- 18.Barsky D, Laurence TA & Venclovas Č (2010) How Proteins Slide on DNA In Biophysics of DNA-Protein Interactions (Williams MC & III LJM, eds), pp. 39–68. Springer New York. [Google Scholar]
- 19.Noller HF, Lancaster L, Zhou J & Mohan S (2017) The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol 24, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ & Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W & Rodnina MV (2016) Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol 23, 342–348. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS & Goldman YE (2013) Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol 20, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caliskan N, Katunin VI, Belardinelli R, Peske F & Rodnina MV (2014) Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Petrov A, Tsai A, O’Leary SE & Puglisi JD (2013) Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol 20, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H-K, Liu F, Fei J, Bustamante C, Gonzalez RL & Tinoco I (2014) A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A 111, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Namy O, Moran SJ, Stuart DI, Gilbert RJC & Brierley I (2006) A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin P, Yu D, Zuo X & Cornish PV (2014) Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep 15, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S, Wen J-D, Bustamante C & Tinoco I (2015) Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell 160, 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai VP, Frank F, Righini M, Lee A, Tinoco I & Bustamante CJ (2018) Simultaneous Force and Fluorescence Measurements on Single Ribosomes Demonstrate that mRNA Secondary Structures do not Restrict EF-G Catalyzed Translocation. Biophysical Journal 114, 592a.29414705 [Google Scholar]
- 30.Ermolenko DN & Noller HF (2011) mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol 18, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z & Noller HF (2012) Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A 109, 20391–20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peske F, Savelsbergh A, Katunin VI, Rodnina MV & Wintermeyer W (2004) Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol 343, 1183–1194. [DOI] [PubMed] [Google Scholar]
- 33.Tinoco I & Bustamante C (2002) The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem 101–102, 513–533. [DOI] [PubMed] [Google Scholar]
- 34.Tinoco I (2004) Force as a Useful Variable in Reactions: Unfolding RNA. Annual Review of Biophysics and Biomolecular Structure 33, 363–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrosyan R (2017) Improved approximations for some polymer extension models. Rheol Acta 56, 21–26. [Google Scholar]
- 36.Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C & Turner DH (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic animation of mRNA unwinding at the proximal and distal active sites.


