Abstract
RNA-mediated interference (RNAi) is a recently discovered process by which dsRNA is able to silence specific gene functions. Although initially described in plants, nematodes and Drosophila, the process is currently considered to be an evolutionarily conserved process that is present in the entire eukaryotic kingdom in which its original function was as a defense mechanism against viruses and foreign nucleic acids. Similarly to the silencing of genes by RNAi, viral functions can be also silenced by the same mechanism, through the introduction of specific dsRNA molecules into cells, where they are targeted to essential genes or directly to the viral genome in case RNA viruses, thus arresting viral replication. Since the pioneering work of Elbashir and coworkers, who identified RNAi activity in mammalian cells, many publications have described the inhibition of viruses belonging to most if not all viral families, by targeting and silencing diverse viral genes as well as cell genes that are essential for virus replication. Moreover, virus expression vectors were developed and used as vehicles with which to deliver siRNAs into cells. This review will describe the use of RNAi to inhibit virus replication directly, as well as through the silencing of the appropriate cell functions.
Keywords: Viruses, RNA interference, RNAi, Small interfering RNA, siRNA
Introduction
RNAi activity is a cross-kingdom phenomenon, active in organisms ranging from fungi to humans, by which introduction of dsRNA leads to the silencing of its corresponding matching gene. Prior to the first description of the phenomenon in nematodes a decade ago, gene silencing had been described in plants and in fungi and was termed posttranscriptional gene silencing (PTGS) [1, 2] and quelling [3, 4] respectively. These phenomenon were later found to act similarly to RNAi.
One of the main characteristics of RNAi activity is the processing of the dsRNA molecule into small 19- to 27-nt dsRNA fragments, designated small interfering RNA (siRNA). This is done by the Dicer enzyme, which belongs to the RNase III family [5–8]. Dicer enzymes are multi-domain proteins, about 200 kDa in size, which possess an RNA helicase domain, a PAZ domain for binding nucleic acids, preferably to RNA [8, 9], a catalytic domain and a double-strand-RNA-binding domain (dsRBD) [10]. For its nuclease activity the enzyme recognizes the ends of the molecule and excises dsRNA fragments from one end to form 19- to 27-nt-long siRNA molecules with a 5′ phosphate end and an overhang of 3′ dinucleotides [11, 12]. Interestingly, E. coli RNase III can be used to perform the dsRNA cleavage to a fully active siRNA substrate that can be used as a siRNAs pool for gene silencing [13, 14].
Consequently, the dsRNA molecules unwind in an ATP-dependent process, and one of the strands is incorporated into the RNA interference silencing complex (RISC). The complex is guided by the single-strand siRNA to the complementary target RNA, which is then cleaved resulting in gene silencing (Fig. 1).
Fig. 1.
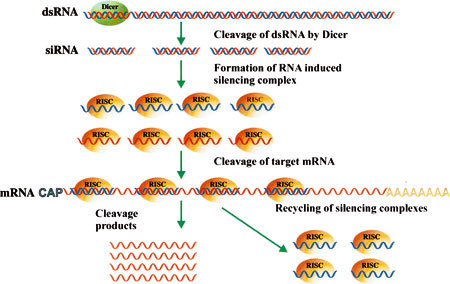
Schematic representation of steps and mechanisms leading to gene silencing by the RNAi process
The main, if not the only protein that constitutes the RISC complex is Argonaute 2 (Ago-2) [15–18], which is characterized by the presence of two domains, one of which is PAZ that is also found in some Dicers [17, 19, 20]. The PAZ domains of both the Dicer and the Ago-2 proteins interact with the 3′ end of the siRNA [21, 22], with which they thereby form a three-component connection [23, 24]. This three-component interaction, mediated by the PAZ domains, is probably responsible for the transfer of newly formed siRNA to RISC [23]. The second domain that characterizes Ago-2 is a PIWI domain that is structurally similar to the RNasHI and RNaseHII domains [23–25]. Furthermore, a bioinformatics approach revealed that the PIWI domain from Drosophila melanogaster is related to the endonuclease V protein family [15]. All these findings indicate that Ago-2 exhibits a complete set of RISC features (Fig. 2).
Fig. 2.
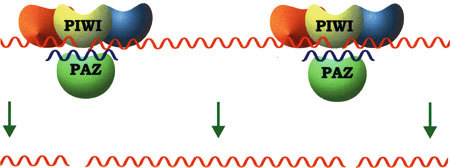
The final RNA cleavage by the RISC apparatus
Induction of the interferon response of the dsRNA by activating the dsRNAi-dependent protein kinase, in particular, PKR, renders RNAi ineffective for gene silencing in interferon-producing organisms [26, 27]. The pioneering work by Elbashir et al. [11, 12] was a breakthrough in the held, in that it showed for the first time that the introduction of siRNA into cells can induce gene silencing by the RNAi mechanism; these publications were followed by an outburst of reports of studies that demonstrated that siRNA can silence large numbers of genes in cells of vertebrate origin, as well as viruses.
siRNA can be introduced into cells either by lipofection [28–30] or as part of an expression vector in which the siRNA gene is under the control of a pol III type promoter such as U6 or HI [31–33]. In this form the positive siRNA strand is proceeds by 4–9 bases and the negative siRNA strand followed by 4 or more T’s serving as the transcription terminator. Upon transcription the transcribed RNA is forming a stem loop structure with is than processed by Dicer to produce fully active siRNA (Fig. 1). Based on the same technique the siRNA cassette can be aplied in viral vectors such as Adeno associate, [34, 35] and retro viruses [36–38].
This review will summarize the available data concerning the silencing of viral replication through direct interference with viral genes as well as by the silencing of cell functions involved virus replication.
RNA viruses
Picorna viruses
Picorna viruses are nonenveloped viruses with a genome of positively oriented single-strand (ss)RNA, 7–8.5 kb in size. The best-known virus of this family is the polio virus, which causes poliomyelitis and that is still a threat in several developing countries. The genome carries one open reading frame and the newly synthesized polyprotein is cleaved by an autoproteolytic process into three polypeptides—P1–P3—which are than cleaved into mature proteins. P1 polypeptide carries the viral structural genes VP 1–4 as well as La protease, whereas P2 and P3 carry the non-structural proteins (NSPs) of the virus.
One of the first reports of the potential of siRNA to inhibit animal virus replication was that by Gitlin et a1. [39], concerning the polio virus. To show viral inhibition two siRNAs were tested: one targeted to the viral protein 3 (VP3) gene, and the second designed to cleave the polymerase gene (3D), which is a non-structural protein. In both cases there was almost complete inhibition as indicated by the reduction of virus titers and by significant decreases of viral RNA and protein in treated cells.
One cardinal issue in the use of RNAi as a therapeutic approach is that of viruses that escape as a result of the selective pressure on the target sequences to mutate, which suppresses the silencing activity of the siRNA molecule. This phenomenon is exactly what was recorded when the inhibition of poliovirus was examined over a long period of time: even a single mutation resulted in G:U mismatching that suppressed the viral inhibition by the siRNA. Moreover, in each experiment several escaping viruses were isolated [40] that exhibited different mutations at the siRNA site. To overcome this adverse effect, Gitlin et al. [41] used a mixture of siRNA molecules originated from a Dicer-treated of 1-kbp segment dsRNA that corresponded to the viral sequences. Since the mixture contained siRNA molecules aimed against numerous sites on the viral genome, a small number of mutations should not affect the overall inhibitory effect and would not permit the emergence of revertantes. When the experiments were performed no escaping viruses could be detected, at least for the duration of the experiments.
A similar but not identical approach was taken by Kahana et al. [41], in order to inhibit replication of the foot-and-mouth disease virus (FMDV). FMDV also belongs to the Aphtopicornavirus genus of the Picornaviridae family infecting cloven-hoofed animals and is the most important farm animal pathogen. In order to silence the virus completely Kahana et al. [41] searched among all the viral entries in the GenBank database for viral sequences that showed 100% homology with any FMD virus over at least a 22-base-long segment. The analysis revealed three such sequences, which became the bases for synthesizing three siRNA molecules corresponding to these three sites. When cells were transfected with individual siRNA molecules prior to being infected, inhibition rates were between 80 and 90%; when they were simultaneously transfected with all three, complete inhibition of virus replication was obtained, as observed by the use of real-time RT-quantitive (q)PCR and virus titers. It could be argued that this approach would also lead to inhibition of all the seven different viral serotypes through use of the same siRNAs molecules; and this should be achievable since the target sequences should be present in most if not all FMDVs, regardless of their origin. Moreover, the conservation of sequences throughout viral evolution indicates that there is a very strong selective pressure against mutations, which has preserved the sequences from changes in the course of virus evolution. This same selective pressure would maintain the siRNAs target sequences unchanged, so ensuring the proper activity of the described siRNAs [41].
As with other viruses, targeted siRNA sequences can be found throughout the genome. In the case of FMDV, siRNA targeted to the VP1 gene, which is the least conserved gene among all FMDV virus genes, was also used successfully to inhibit virus replication in cells; it also reduced the virus titers of infected suckling mice that received a subcutaneous injection of the siRNA-expressing vector [42].
Hepatitis C
Hepatitis C is an enveloped virus and is the sole member of the genus Hepacvirus of the Flaviviridae family. The viral genome is a positively oriented single-stranded RNA, about 9.6 kb in length. It carries a single open reading frame and the translated 3000-aa polyprotein is cleaved by host as well as viral proteinases, to produce 10 mature proteins. The N terminus of the genome encodes for the core, E1 and E2 structural proteins and the remainder of the genome encodes for the non-structural ones. The 5′ untranslated region (5′ UTR) carries the transcription initiation site for the transcription of the positive-strand RNA, and an internal ribosome entry site (IRES) for the initiation of viral protein synthesis.
Infection with the HPC virus is one of the major viral diseases of humans worlwide, and it is estimated that about 170,000,000 people are HPC seropositive [43, 44]. Obviously it was one of the first viruses to be checked for the possibility of inhibition by means of RNAi.
One of the main approaches to the design of siRNA targets is the identification of highly conserved regions among isolates of the specific virus, as was described for FMDV [45]. In HPCV, the most conserved region is the 5′ UTR, particularly the IRES region. Yokota and coworkers [46] used this design strategy to design siRNA molecules targeted to various regions of the 5′ UTR IRES sequence. In tests of six siRNAs genes, silencing levels were not uniform: only the siRNA corresponding to a region upstream of the AUG start codon could provide a high inhibition rate. Not only were all the genes not equally effective; there was an siRNA molecule that had an opposite effect, i.e., it increased the replication rate [46]. Contrary to this observation Takigawa et al. [47] reported that an siRNA targeted to this region was unable to inhibit the replication of the virus. On the other hand, the same authors found that siRNA targeted against the NS3 and NS5B regions, which are less conserved than the 5′ UTR, were also highly effective in inhibiting virus activity and achieved 90% inhibition [47].
Influenza virus
The influenza virus is a member of the Orthomyxoviridae and is responsible for acute upper respiratory disease. The viral genome is composed of eight negatively oriented ssRNA segments.
Influenza virus infection is a severe health problem worldwide, causing millions of cases of disease and an estimated 500,000 deaths each year. Resorting and recombination events involving avian influenza virus genomes could produce a new and highly virulent virus, able to cross the species barrier from birds to humans, with a high mortality rate among infected individuals [48–50].
As there are eight genome segments, siRNA molecules targeted to each of the different genomes were tested, and several siRNA molecules that were targeted to PA, PB1 and NP segments were found to have the most profound effect on virus replication in MBDK cells [51]. This result does not imply that siRNAs targeted to other genes would be less effective, as was shown by Hui et al. [52]. It was shown that an siRNA targeted against the virus matrix (MI) gene effectively inhibited virus replication in 293T cells [52].
Not only could it be shown that siRNA effectively inhibited influenza replication in vitro, but the same approach led to inhibition of virus replication in mice in vivo. When siRNA molecules were prepared in a complex with Oligofectamine or polyethylenimine (PEI) and delivered intravenously or intranasally [53] there was a marked reduction in virus replication. When siRNA was targeted to NP and PA genes simultaneously, there was a broad range of reductions of replication of H5, H7 and H9 viral serotypes. The most important result was the inhibitory effect of the siRNA on the highly pathogenic avian influenza viruses from South to East Asia that are capable of infecting humans [54]. Not only did this approach succeed in vivo, when double-stranded siRNA was introduced into mice, but the siRNA was also highly active when delivered as a gene under a Pol III type promoter in a lentivirus vector [54].
Respiratory syndrome associated coronavirus (SARS-CoV)
SARS-CoV, a novel and highly dangerous coronavirus was identified in 2003. It was originally an avian virus that crossed the species barrier to infect humans, and is characterized by its high mortality rate among infected people. This phenomenon caused a severe health problem worldwide, and especially in Southeast Asia and Canada, where cases of human infection were found. The genome, like other coronaviruses is a ssRNA of positive orientation, about 30 kb in length. As in the cases of other described viruses, several siRNAs were targeted to various locations on the viral genome, against the viral replicase [55] and the spike gene [56–58] were designed; their ability to silence the expression of those genes, and also to inhibit viral replication was demonstrated.
HIV
As HIV presents a major health problem that has reached pandemic proportions, especially in the developing countries [59], and it was one of the first viruses to be tested for the ability of RNAi approach to be inhibited [60, 61]. Since several excellent reviews on this subject have been published, only one aspect of virus inhibition will be discussed in the present review, namely the various means that were used to overcome the phenomenon of virus escapes that resulted from the high rate of mutation of the virus and prevented the future use of this approach for therapeutic purposes. Because of the exceedingly high mutation rate of viral genome, the siRNA target sequences are also mutated at a very high rate [62, 63], which impairs the long-term effectiveness of siRNAs, and eventually leads to the emergence of escaping viruses that overcome the effect of siRNA on virus replication [64–66]. The severity of the phenomenon is indicated by the facts that when the sequences of siRNA targets on the genomes of the virus were determined it was found that not only mutations at the target sequences could break down siRNA activity, but that mutations that are adjacent to the actual target sequences could also stop siRNA activity.
Several attempts were made to address this issue. One approach involved the scanning of virus sequences in order to find regions of conserved sequences that were located across the viral genome and raising siRNA genes corresponding to these regions [67]. By means of this approach, siRNA molecules corresponding to gag, pol, int and vpu sequences were shown to inhibit three different HIV strains effectively [67]. Although the long-term protection was not evaluated it seems that this approach has the potential for finding viral sequences in which a mutation would result in the production of attenuated or non-viable viruses. Moreover, the use of several such siRNA genes simultaneously would have an excellent chance to protect against the appearance of escape viruses, by overcoming the selective pressure against siRNA activity, as predicted by a model developed by Leonard and Shaffer [68]. This is a stochastic computer model for testing different approaches to overcome the appearance of mutations that escape from siRNA; it predicted that the use of several siRNAs targeted to different locations would dramatically increase siRNA efficacy. The model also predicted that a small increase in the number of siRNA targets would drastically increase the efficiency of the siRNA approach, by reducing or eliminating the appearance of escaping viruses. That study [68] also highlighted the importance of the transformation efficiency of the siRNA gene or molecules in ensuring that each infected cell contain the siRNA molecules; if there were HIV-producing cells that did not carry the inhibitory molecules, virus replication would continue in these cells and so increase the likelihood of the appearance of escaping viruses.
Another approach to the inhibition of HIV replication involves directing siRNAs against cell functions indispensable to the virus life cycle. Such functions are performed by cell viral receptors on the cell membrane, such as CD4, CXCR4, and CCR5. When siRNA targeted to the CD4, CXCR4, and CCR5 genes were tested it was found that HIV replication could be suppressed through the repression of virus entry, which conferred resistance to HIV-1 infection [69]. An interesting approach that was taken in the same study [69] involved the use of bispecific short hairpin (sh)RNA molecules that were targeted against two different genes. Even though silencing of CD4 can suppress virus infection effectively, such silencing would not be a practicable approach because of the importance of the CD4 protein in the immune system.
A gene that would be a more realistic target is CCR5, the major HIV-1 co-receptor in macrophages. This molecule is dispensable, since CCR5−/− homozygote individuals are protected against viral infection without any abnormal phenotype.
Transfection of a specific CCR5 siRNA into macrophage, following virus infection, reduced virus replication almost completely, but in primary human microglia at 15 days post challenge, inhibition was not complete [70]. Another approach to the complete inhibition of HIV-1 replication involved the targeting of two sites: one being an entry way such as the CCR5 gene; a second being a viral gene such as the p24 gene. When this approach was tested using siRNAs against both CCR5 and viral p24 simultaneously [71] virus reproduction was completely eliminated, even when the test was applied 15 days post challenge. This experiment demonstrated that inhibition of conserved virus functions together with that of cellular functions important to the virus life cycle can have a sustained inhibitory effect on HIV-1 replication and can be pursued for therapeutic purposes.
A different cell functional function that was tested was poly(ADP-ribose) polymerase 1 (PARP-1), which is a nuclear enzyme that catalyses the transfer of NAD+ ADP-ribose moiety to nuclear proteins including itself. PARR-1 is needed for the activation of NF-κB-dependent target genes, as the human immunodeficiency virus type 1(HIV-1) long-terminal repeat (LTR). Lack of PARP-1 prevents the integration of HIV-1 into the genome, as was shown in PARP-1 knockout cells, which makes it an excellent choice for effectively suppressing HIV-1 replication. As predicted, siRNAs targeted to the PARP-1 gene efficiently suppressed the replication of HIV-1 in both human HeLa and J111 cell lines. Furthermore, silencing the gene effectively restricted virus integration into the genome [72].
DNA viruses
Hepatitis B virus
Hepatitis B (HB) is a major health problem in many countries, and it is estimated that about 400 million people worldwide are carriers. This virus infection can cause liver cirrhosis and hepatocellular carcinoma, which is responsible for about 1 million fatalities annually. The virus genome is a partially double-stranded small DNA that has a relaxed-circular DNA structure (Fig. 3). One of the main unique features of its genome DNA replication is that it has a reverse transcription step [73].
Fig. 3.
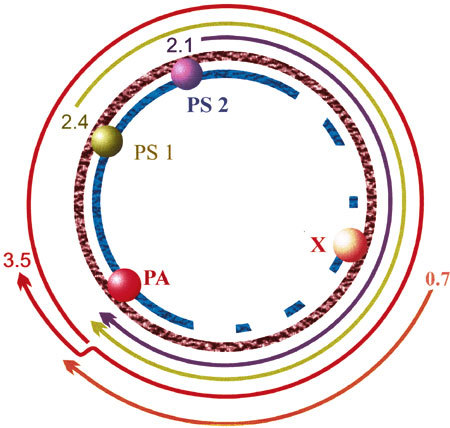
The genome and transcripts of hepatitis B virus. PA, PS1, PS2 and X are the viral promoters
In contrast to RNA viruses, in which siRNA is also able to cleave their genome, in DNA viruses the inhibition is due to silencing of viral functions through viral mRRNA cleavage, similarly to its activity on cellular functions.
As was described with regard to other viruses, siRNA targeted against a specific site can inhibit virus replication. Several transit expressed siRNAs targeted to sequences located throughout the viral genome were tested and were found to elicit large-scale inhibition of virus replication [74]. As mentioned earlier, one of the main concerns regarding siRNA delivery is transfection efficiency, since siRNA molecules, either in the form of dsRNA or as expressible vectors, must enter all infected cells in order to stop virus replication completely. One way to ensure this is through the use of stable transformed cells, which guarantees the presence of siRNA in every infected cell, as reported by Chen et al. [75], who tested 10 different siRNA genes aimed at all viral genes and were able to show that viral RNA levels could be reduced by up to 90%. One important aspect of using siRNA activity against DNA viruses is the need to show the inhibition of viral DNA replication, and when the amounts of viral DNA were quantified almost no viral DNA could be detected, which demonstrated the inhibition of genome replication similarly to the inhibition of the genome replication in RNA viruses [75]. Since the result of an in vitro assay is only an indicator for a practical application the potential of this approach must be tested in vivo, as was performed by Giladi et al. [76] and Uprichard et al. [77]. Giladi et al. [76] showed that co-injection of a plasmid carrying an HBV genome and a plasmid carrying siRNA genes corresponding to sequences of the small HBV surface antigene (HbsAg) there was a marked decrease of both HBsAg and HBeAg, and a 50% decrease of all three viral mRNAs. By far the most dramatic effect was on viral DNA replication: up to 3 days after injection the viral DNA remained at undetectable levels.
Uprichard et al. [77] used a somewhat a different system to achieve similar results: infection of HBV transgenic mice by an Adenovirus vector carrying siRNA genes targeted to the 2.1, 2.4 and 3.5-kb overlapping sequences was able to reduce the levels of HBsAg and HBeAg in the serum of infected mice. Silencing of the 2.1 and 3.5-kb viral RNA was maintained for atleast 20 days post infection, which confirmed the effectiveness of this approach. At day 17 post infection DNA replication remained at undetectable levels, and remained so until the end of the experiment on day 26.
Herpes viruses
Herpes viruses are one of the largest groups of DNA viruses, and they infect most if not all vertebrates. The genome is dsDNA, ranging from 90 to 150 kbp in length, and the virus replicates in the nucleus. Little work has been published on the use of siRNA to inhibit herpes viruses. Bhuyar et al. [78] demonstrated the ability of siRNA to silence the gE glycoprotein, a transmembrane glycoprotein involved in cell-to-cell movement.
Conclusions
In summary, RNAi and its employment in virology offer for the first time the possibility of controlling viral diseases. This approach has a similar potential to impact on human, animal and plant health to that of antibiotics in the last century.
References
- 1.Ingelbrecht I., Van Houdt H., Van Montagu M., Depicker A. Proc. Natl. Acad. Sci. USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baulcombe D.C. Plant Mol. Biol. 1996;32:79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 3.Romano N., Macino G. Mol. Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 4.Cogoni C., Romano N., Macino G. Antonie Van Leeuwenhoek. 1994;65:205–209. doi: 10.1007/BF00871948. [DOI] [PubMed] [Google Scholar]
- 5.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 6.Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Carmell M.A., Hannon G.J. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 9.G.J. Hannon, P.D. Zamore, in RNAi: A Guide to Gene Silencing, ed. by G.J. Hannon (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2003) pp 87–108
- 10.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir S.M., Lendeckel W., Tuschl T. Genes Dev. 2001a;15:185–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbashir S.M., Martinez J. EMBO J. 2001b;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. Proc. Natl. Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang D., Goga A., Bishop J.M. Meth. Mol. Biol. 2004;252:471–482. doi: 10.1385/1-59259-746-7:471. [DOI] [PubMed] [Google Scholar]
- 15.Rand T.A., Ginalski K., Grishin N.V., Wang X. Proc. Natl. Acad. Sci. USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 17.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 18.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Lingel A., Simon B., Izaurralde E., Sattler M. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 20.Lingel A., Simon B., Izaurralde E., Sattler M. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 21.Ma J.B., Ye K., Patel D.J. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson R.H., Nicholson A.W. Genome. 2002;13:67–73. doi: 10.1007/s00335-001-2119-6. [DOI] [PubMed] [Google Scholar]
- 23.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 24.Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 25.Parker J.S., Roe S.M., Barford D. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel R.C., Sen G.C. Cell Mol. Biol. Res. 1994;40:671–682. [PubMed] [Google Scholar]
- 27.Katze M.G. Trends Microbiol. 1995;3:75–78. doi: 10.1016/S0966-842X(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 28.Sioud M., Sorensen D.R. Biochem. Biophys. Res. Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Shan P., Jiang D., Noble P.W., Abraham N.G., Kappas A., Lee P.J. J. Biol. Chem. 2004;279:10677–10684. doi: 10.1074/jbc.M312941200. [DOI] [PubMed] [Google Scholar]
- 30.Liu C., Shi Y., Han Z., Pan Y., Liu N., Han S., Chen Y., Lan M., Qiao T., Fan D. Biochem. Biophys. Res. Commun. 2003;312:780–786. doi: 10.1016/j.bbrc.2003.10.186. [DOI] [PubMed] [Google Scholar]
- 31.Futami T., Miyagishi M., Seki M., Taira K. Nucl. Acids Res. Suppl. 2002;2:251–252. doi: 10.1093/nass/2.1.251. [DOI] [PubMed] [Google Scholar]
- 32.Matsukura S., Jones P.A., Takai D. Nucl. Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Wetering M., Oving I., Muncan V., Pon Fong M.T., Brantjes H., van Leenen D., Holstege F.C., Brummelkamp T.R., Agami R., Clevers H. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinkenburg O., Platz J., Beisswenger C., Vogelmeier C., Bals R. J. Virol. Meth. 2004;120:119–122. doi: 10.1016/j.jviromet.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Vargas I., Travanty E.A., Keene K.M., Franz A.W., Beaty B.J., Blair C.D., Olson K.E. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 36.An D.S., Xie Y., Mao S.H., Morizono K., Kung S.K., Chen I.S. Hum. Gene Ther. 2003;14:1207–1212. doi: 10.1089/104303403322168037. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.S., Hmama Z., Mui A., Reiner N.E. J. Biol. Chem. 2004;279:9379–9388. doi: 10.1074/jbc.M310638200. [DOI] [PubMed] [Google Scholar]
- 38.Li M.J., Rossi J.J. Meth. Enzymol. 2005;392:218–226. doi: 10.1016/S0076-6879(04)92013-7. [DOI] [PubMed] [Google Scholar]
- 39.Gitlin L., Karelsky S., Andino R. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 40.Gitlin L., Stone J.K., Andino R. J. Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahana R., Kuznetzova L., Rogel A., Shemesh M., Hai D., Yadin H., Stram Y. J. Gen. Virol. 2004;85:3213–3217. doi: 10.1099/vir.0.80133-0. [DOI] [PubMed] [Google Scholar]
- 42.Chen W., Yan W., Du Q., Fei L., Liu M., Ni Z., Sheng Z., Zheng Z. J. Virol. 2004;78:6900–6907. doi: 10.1128/JVI.78.13.6900-6907.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg S. J. Mol. Biol. 2001;313:451–464. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 44.Thomson B.J., Finch R.G. Clin. Microbial Infect. 2005;11:86–94. doi: 10.1111/j.1469-0691.2004.01061.x. [DOI] [PubMed] [Google Scholar]
- 45.Kronke J., Kittler R., Buchholz F., Windisch M.P., Pietschmann T., Bartenschlager R., Frese M. J. Virol. 2004;178:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokota T., Sakamoto N., Enomoto N., Tanabe Y., Miyagishi M., Maekawa S., Yi L., Kurosaki M., Taira K., Watanabe M., Mizusawa H. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takigawa Y., Nagano-Fujii M., Deng L., Hidajat R., Tanaka M., Mizuta H., Hotta H. Microbiol. Immunol. 2004;48:591–598. doi: 10.1111/j.1348-0421.2004.tb03556.x. [DOI] [PubMed] [Google Scholar]
- 48.Xu X., Subbarao K., Cox N.J., Guo Y. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 49.Class E.C., Osterhaus A.D., van Beek R., De Jong J.C., Rimmelzwaan G.F., Senne D.A., Krauss S., Shortridge K.F., Webster R.G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 50.Guan Y., Poon L.L., Cheung C.Y., Ellis T.M., Lim W., Lipatov A.S., Chan K.H, Sturm-Ramirez K.M., Cheung C.L., Leung Y.H., Yuen K.Y., Webster R.G., Peiris J.S. Proc. Natl. Acad. Sci. USA. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ge Q., McManus M.T., Nguyen T., Shen C.H., Sharp P.A., Eisen H.N., Chen J. Proc. Natl. Acad. Sci. USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hui E.K., Yap E.M., An D.S., Chen I.S., Nayak D.P. J. Gen. Virol. 2004;85:1877–1884. doi: 10.1099/vir.0.79906-0. [DOI] [PubMed] [Google Scholar]
- 53.Ge Q., Filip L., Bai A., Nguyen T., Eisen H.N., Chen J. Proc. Natl. Acad. Sci. USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tompkins S.M., Lo C.Y., Tumpey T.M., Epstein S.L. Proc. Natl. Acad. Sci. USA. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J., Chen Y.G. J. Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X., Wang Y., Ning H., Zhang S., Chen W., Babiuk L.A., Chang Z. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu C.J., Huang H.W., Liu C.Y., Hong C.P., Chan Y.L. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin Z.L., Zhao P., Zhang X.L., Yu J.G., Cao M.M., Zhao L.J., Luan J., Qi Z.T. Biochem. Biophys. Res. Commun. 2004;324:1186–1193. doi: 10.1016/j.bbrc.2004.09.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogunbodede E.Q. Intl. Dent. J. 2004;54:352–360. [PubMed] [Google Scholar]
- 60.Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 61.Jacque J.M., Triques K., Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coffin J.M. Cell. 1986;46:1–4. doi: 10.1016/0092-8674(86)90851-2. [DOI] [PubMed] [Google Scholar]
- 63.Roberts J.D., Bebenek K., Kunkel T.A. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 64.Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. J. Virol. 2003;77:1153–1155. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das A.T., Brummelkamp T.R., Westerhout E.M., Vink M., Madiredjo M., Bernards R., Berkhout B. J. Virol. 2004;178:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westerhout E.M., Ooms M., Vink M., Das A.T., Berkhout B. Nucl. Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.L.J. Chang, X. Liu, J. He, Gene Ther. Mar 3 (2005) [DOI] [PubMed]
- 68.Leonard J.N., Schaffer D.V. J. Virol. 2005;179:1645–1654. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson J., Banerjea A., Akkina R. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 70.Cordelier P., Morse B, Strayer D.S. Oligonucleotides. 2003;13:281–294. doi: 10.1089/154545703322616961. [DOI] [PubMed] [Google Scholar]
- 71.Song E., Lee S.K., Dykxhoorn D.M., Novina C., Zhang D., Crawford K., Cerny J., Sharp P.A., Lieberman J., Manjunath N., Shankar P. J. Virol. 2003;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kameoka M., Nukuzuma S., Itaya A., Tanaka Y., Ota K., Ikuta K., Yoshihara K. J. Virol. 2004;78:8931–8934. doi: 10.1128/JVI.78.16.8931-8934.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Locarnini S. Semin Liver Dis. 2004;24(Suppl. 1):3–10. doi: 10.1055/s-2004-828672. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X.N., Xiong W., Wang J.D., Hu KW., Xiang L., Yuan Z.H. World J. Gastroenterol. 2004;10:2967–2971. doi: 10.3748/wjg.v10.i20.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y., Du D., Wu J., Chan C.P., Tan Y., Kung H.F., He M.L. Biochem. Biophys. Res. Commun. 2003;311:398–404. doi: 10.1016/j.bbrc.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 76.Giladi H., Ketzinel-Gilad M., Rivkin L., Felig Y., Nussbaum O., Galun E. Mol. Ther. 2003;8:769–776. doi: 10.1016/S1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 77.Uprichard S.L., Boyd B., Althage A., Chisari F.V. Proc. Natl. Acad. Sci. USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhuyar P.K., Kariko K., Capodici J., Lubinski J., Hook L.M., Friedman H.M., Weissman D. J. Virol. 2004;78:10276–10281. doi: 10.1128/JVI.78.19.10276-10281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


