Abstract
Porcine epidemic diarrhea virus (PEDV), which causes 80–100% mortality in neonatal piglets, is one of the most devastating viral diseases affecting swine worldwide. To date, the lack of effective vaccines and drugs is the main problem preventing control of the global spread of PEDV. In this study, we produced PEDV virus-like particles (VLPs) composed of S, M, and E proteins with a baculovirus expression system and tested them via indirect immunofluorescence assay (IFA)and Western blot analysis. Electron microscopy showed that the morphological structure of the PEDV VLPs was similar to that of the protovirus. Microneutralization assays and ELISpot analysis demonstrated that PEDV VLPs induced highly specific antibody responses and Th2-mediated humoral immunity. As a result, the PEDV VLPs displayed excellent immunogenicity in mice. Therefore, a VLP-based vaccine has the potential to prevent PEDV infection.
Keywords: Porcine epidemic diarrhea virus, Virus-like particles, Neutralizing antibody, Immune response
Introduction
Porcine epidemic diarrhea virus (PEDV) belongs to the genus Alphacoronavirus in the family Coronavirus. PEDV is spherical and has a diameter of approximately 130 nm [1]. PEDV is a single-stranded positive-sense RNA genome with a size of approximately 28 kb that encodes four separate structural proteins: the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. The S protein is a vital protein for regulating the recognition and interaction between specific cell receptors and the virus as well as for inducing neutralizing antibody generation. The M protein, however, is the essential component for viral assembly [2, 3]. The E protein likely facilitates the release of the assembled virions. The presence of both the E protein and the M protein is sufficient for the formation and release virus-like particles [4]. The N protein is essential for viral transcription but is not necessary for the formation of virus-like particles [5].
Porcine epidemic diarrhea (PED) was first reported in Europe in the early 1970 sand has since caused great economic losses for the swine industry in Asia [6–10]. It has become an endemic epizootic in Asia in recent decades and causes 80–100% mortality in neonatal piglets. However, the disease had never occurred in American countries until 2013. PEDV emerged in the United States in 2013 and has since swept across North America, including Canada and Mexico, resulting in the deaths of more than 8 million newborn piglets in the United States alone during that 1-year epidemic period [11–14]. PEDV is one of the most devastating viral diseases affecting swine in the world and has caused significant financial effects on the global pork industry.
There are no effective PED vaccines or drugs, and some veterinarians have suggested that feedback material is a fundamental strategy to control PED during epidemic or endemic outbreaks. One of the most important measures for controlling PED outbreaks is vaccination. To date, there are the China CV777-attenuated or inactivated vaccines, the Japanese P-5 V live vaccine, the Korea SM98-1 live or killed vaccine, and the DR-13 oral live vaccine [3, 15, 16].
Recent vaccine candidates have been engineered to be replication-deficient or a virulent, which has eliminated the risks associated with live-attenuated vaccines regaining virulence. Other PEDV candidate vaccines that are currently being developed include spike (S) protein nanoparticles [17] and a modified tobacco mosaic virus(TMV)based on the full-length S DNA and S1 subunit protein [18], which both induce neutralizing antibodies against PEDV. Strong cellular and humoral immune responses are elicited by these vaccines, which can be administered orally. However, there is existing immunity against the viral vector. The inactivated vaccine is safe, and the production procedures are simple, but this vaccine only elicits a Th2-bias immune response and requires a specific adjuvant. The live-attenuated vaccine can induce strong cellular and humoral immune responses, but there is a risk of virulence reversion [19]. Virus-like particles (VLPs) are composed of one or more viral structural proteins, and their morphologies closely resemble the native virus. VLPs have no risks of reversion to virulence and can elicit robust immune responses compared with inactivated or live-attenuated virus vaccines [20, 21]. Furthermore, VLPs can be generated through artificial methods for any pathogen.
It has been reported that the MERS-CoV S protein is essential for inducing neutralizing antibodies, but the S and M proteins alone cannot assemble into VLPs [22, 23]. The M protein is necessary for VLP composition, and the E protein is important for VLP assembly [24]. Thus, the PEDV S, M, and E proteins are necessary for VLP assembly via co-expression in Sf9 cells.
In this study, we used a baculovirus expression system to successfully construct a recombinant baculovirus containing the spike (S), membrane (M), and envelope (E) protein genes. We obtained self-assembling VLPs by infecting Sf9 cells with this recombinant baculovirus. Then, we demonstrated the structural integrity of the VLPs and evaluated their immunogenicity in mice. Our results have established the foundation for the development of effective vaccines against PEDV.
Materials and methods
Viruses and cells
Vero cells were cultured in DMEM (Gibco, Grand Island, USA) in a culture bottle at 37 °C and 5% CO2. Sf9 cells were cultured with TNM-FH medium (Sigma, New York, USA) in plates at 27 °C. PEDV was a gift from Professor Hu Guixue from Jilin Agricultural University.
Construction of the recombinant plasmid and indirect immunofluorescence assay (IFA)
The epidemic isolate(AH2012 strain) was chosen, and the PEDV full-length S, M and E genes were optimized for Sf9 cell expression by Sangon Biotech. The optimized S, M, and E genes were cloned into the modified pFastBac Dual plasmid. Then, the recombinant plasmid was transformed into DH10Bac. The recombinant bacmid was then obtained by blue-white spot screening as previously described [22].
An indirect immunofluorescence assay was performed to determine whether the desired proteins were expressed in the Sf9 cells infected with recombinant baculovirus. This method has been described previously [22]. Briefly, the infected Sf9 cells were cultured at 27 °C for 48 h and then fixed with pre-cooled 4% paraformaldehyde for 30 min. Then, the cells were washed three times with PBST. Subsequently, the cells were incubated with mouse polyclonal antibodies against the S, M, and E PEDV proteins for 1 h. Next, the cells were washed three times with PBST. Finally, the cells were incubated with FITC-labeled goat antibody (Sigma, Ronkonkoma, New York, USA) against mouse IgG containing 0.3% Evans blue at 37 °C for 1 h. The cells were observed with a fluorescence microscope after three washes with PBST.
Production and purification of VLPs
The constructed recombinant Bacmid containing the S, M, and E protein genes was transfected into Sf9 cells. At 96-h post-infection (hpi), the desired recombinant baculovirus was harvested, and the baculoviral titers were determined using a BacPak Baculovirus Rapid Titer Kit (Clontech, California, USA) for determining the inoculation dose when the VLPs are produced by cultivation in large quantities. Sf9 cells were infected with recombinant baculovirus at 96hpi, after which the PEDV VLPs were harvested. The supernatants were cleared of cell debris by centrifugation at 30,000×g for 1 h. Then, the supernatants crossed a 30–50% discontinuous sucrose gradient to isolate the VLPs; this procedure was previously described [25].
Western blot analysis
To further evaluate PEDV VLP expression, we performed a modified Western blot assay [26]. The harvested VLPs were boiled for 10 min and then separated via SDS-PAGE. Then, the VLPs in the gel were transferred to a pure nitrocellulose blotting (NC) membrane (GE Healthcare Life Sciences, Kent, UK) for 1 h at 20 V. The membrane was blocked in blocking buffer (Thermo, Massachusetts, USA) for 90 min at room temperature. Next, anti-S, -M, and -E mouse polyclonal antibodies were incubated with the membrane for 18 h at 4 °C. The membrane was then washed three times with PBST and incubated with goat anti-mouse IgG-conjugated HRP (Sigma, Ronkonkoma, New York, USA) for 1 h, after which it was washed three times, and protein expression was visualized using an ECL system (Thermo, Massachusetts, USA).
Electron microscopy
PEDV VLPs were coactivated with 1% sodium phosphotungstate for 5 min. Superfluity was pruned away with filter paper, and the samples were then examined using transmission electron microscopy.
Immunization studies
Six-week-old female BALB/c mice were purchased from the Changchun Animal Breeding Center for Medical Research (Changchun, China). The mice were randomly separated into three groups(10 mice/group). One group was immunized with 25 μg purified VLPs per mouse by intramuscular injection at two-week intervals, the second group with PBS alone as the negative control, and the last group with killed PEDV (Fig. 3a).
Fig. 3.
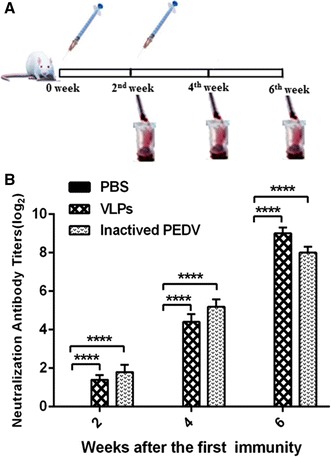
Immunization procedure in mice and the serum neutralizing antibody assay. a Six-week-old mice were vaccinated via intramuscular injection with PBS, VLPs and inactivated PEDV. Blood samples were collected from the retro-orbital plexus puncture every 2nd week after immunization. b Serum-neutralizing anti-PEDV antibody titers. Differences among the groups were analyzed by two-way ANOVA. The titers were delegated on a log2 scale and are shown as the means ± SEM. (**** P < 0.0001)
Microneutralization assay
Blood samples were collected every 2nd week after immunization. The disposal method used for the samples was described previously [22]. The serum levels of specific virus neutralization antibodies (VNA) were examined by serum microneutralization (SN) assays using Vero cells as previously described [27]. Briefly, the serum samples were serially diluted twofold in DMEM without penicillin–streptomycin and serum, and then incubated at 37 °C for 1 h in a 5% CO2 incubator after mixing with 100 TCID50 of the virus stock. These samples were then mixed with Vero cells. After 48 h, the highest serum dilution with no cytopathic effects was determined to be the serum-neutralizing antibody titer.
ELISpot assay
During the second week of the first booster vaccination, red cells were isolated and disrupted, and the splenocytes were successfully separated. After washing twice with RPMI-1640 medium (Gibco, Grand Island, USA)containing 10% fetal bovine serum and 1% penicillin–streptomycin, 5 × 105 splenocytes per well were cultured in RPMI-1640 and simultaneously stimulated with inactivated PEDV in a 96-well ELISpot plate (MABtech, Stockholm, Sweden) at 37 °C in a 5% CO2 incubator for 48 h. The other assay was carried out according to the manufacturer’s instructions. The amount of splenocytes producing IFN-γ and IL-4 was illustrated with spot-forming cells using an ELISpot reader (ELISPOT READER SPECTRUM, AID, Germany) as described previously [28].
Statistical analysis
One-way and two-way ANOVA were used for the statistical analysis. A P value less than 0.01 (P < 0.01) was deemed to display a high degree of statistical significance between the treatment and control groups.
Results
The construction of the recombinant plasmid and the identification of recombinant baculovirus
The modified pFastBac Dual vector containing the PEDV S, M, and E genes was successfully constructed (Fig. 1a), allowing us to obtain the recombinant bacmid.
Fig. 1.
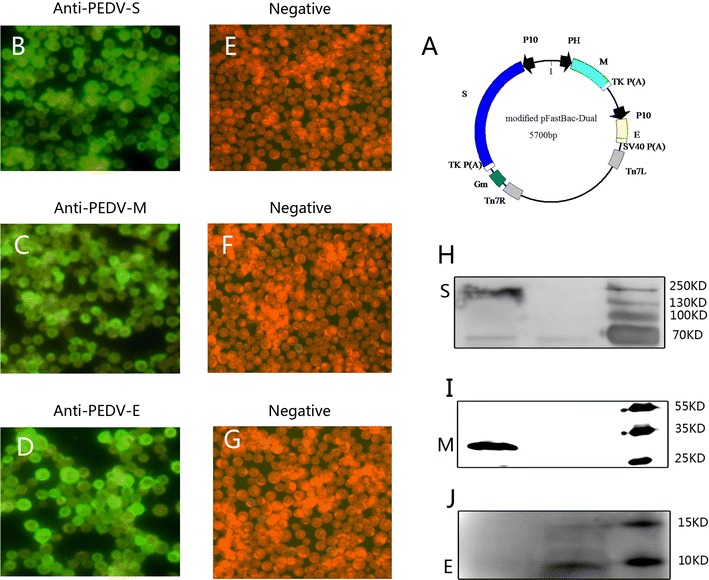
Sketch map of the constructed co-expression plasmid the PEDV S, M and E proteins expressed in Sf9 cells. a The PEDV S, M and E genes were cloned into the different sites in the modified pFastBac-Dual vector. The PEDV S, M and E proteins expressed in Sf9 cells are shown by IFA b–d and e–g are the negative controls. Western blot analysis was used to evaluate the components of the purified VLPs (h–j).
The PEDV S, M, and E genes were expressed in Sf9 cells as shown by indirect immunofluorescence assay(Fig. 1b–d, respectively). Western blot was used to further prove that the VLPs were assembled with S, M, and E proteins. The S, M, and E bands are shown in Fig. 1h–j, respectively.
Morphological observations of PEDV VLP
The recombinant baculovirus expressing the genes of interest was maintained by transfecting Sf9 cells and passaging them three times. The PEDV VLPs were produced by Sf9 cells infected with the recombinant baculovirus, and their morphology was observed by transmission electron microscopy. The purified PEDV VLPs were harvested using inconsecutive sucrose gradient centrifugation for further studies.
Under TEM, the shape and size of the VLPs were similar to those of the naïve virus (Fig. 2a). The shape of the VLPs was similar to a sphere with a diameter of approximately 150 nm; there was also a spike protein around the sphere (Fig. 2b).
Fig. 2.
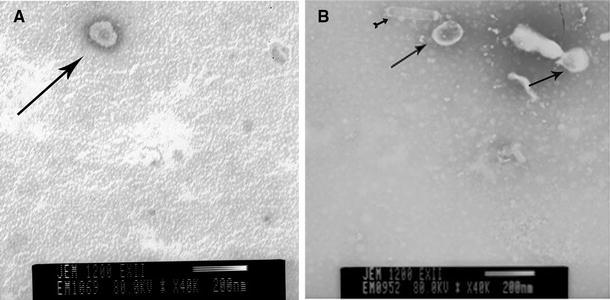
PEDV and PEDV VLPs morphological observations under TEM. a, b The morphologies of the PEDV and PEDV VLPs respectively via TEM
PEDV VLPs induce neutralizing antibodies in mice
The neutralizing anti-PEDV antibody levels in serum from immunized mice were measured. The titers were 3 and 4 two weeks after the first vaccination with VLPs and inactivated PEDV, respectively. The titers were 24 and 54 after the first booster vaccination and 563 and 282 after the second booster vaccination (Fig. 3b). In contrast, neutralizing anti-PEDV antibodies were not detected in the mock groups.
PEDV VLPs significantly enhance Th2-mediated humoral immunity in mice
Changes in IFN-γ and IL-4 levels secreted by CD4+ T cells following vaccination were analyzed using an ELISpot assay. The results showed that secreted IFN-γ levels were not significantly different from those of the control (data not shown). In contrast, the secreted IL-4 levels were significantly different between the two groups (Fig. 4).
Fig. 4.
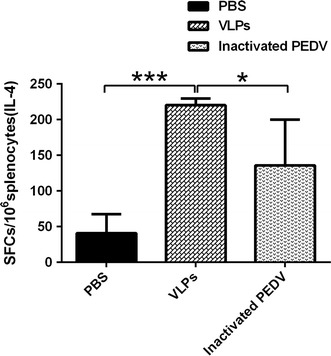
ELISpot assays for IL-4 secretion in splenocytes. PBS is the negative control group, and inactive PEDV and VLPs are the experimental groups. The statistical analysis among groups was performed by one-way ANOVA. A p value less than 0.5 and more than 0.01 was deemed statistically significant (*0.01 < P < 0.5, ***P < 0.001)
Discussion
Due to the PEDV outbreak in the US in 2013, studying PEDV has become a major goal of animal virology scientists. Subsistent commercial vaccines currently cannot prevent infection in piglets [29]. Live-attenuated virus vaccines are often less safe and require strict preservation. Inactivated vaccines are safe for use but induce only very-low-level immune responses. Therefore, the development of new vaccines is necessary.
In our study, a baculovirus expression system was used to produce PEDV VLPs. VLPs structurally and antigenically resemble the virus from which they are derived and can serve as effective stand-alone vaccines or vaccine platforms [30]. To date, several VLP-based vaccines have been produced and commercialized, such as recombinant hepatitis B virus (HBV) [31], human papillomavirus (HPV) vaccine [32], human E virus (HEV) [33], influenza virus [34, 35], and porcine circovirus type 2 (PCV2) [36]. VLPs against RSV [37], norovirus [38], parvovirus B19 [39], and malaria [40] have also entered clinical studies. Many virus VLPs are in different study stages, including VLPs against rabies, hepatitis C virus (HCV), dengue virus, Nipah virus, bovine rotavirus, SARS-CoV, and MERS-CoV [41, 42].
Our preliminary results demonstrated that PEDV VLPs were successfully generated. The VLPs consisted of an eliciting neutralizing antibody S protein and an essential nucleocapsid that contained the M and E proteins. The appearance of the obtained VLPs was similar to that of naïve PEDV. Thus, the VLPs maintained a more perfect immunogenicity than that with other subunit vaccines. Furthermore, the VLPs are more secure than live-attenuated vaccines because they cannot replicate.
PEDV-neutralizing antibody formation is induced by VLPs’ S proteins that contain B-cell-neutralizing epitopes [43, 44]. The M protein contains a conserved linear B-cell epitope that can elicit the formation of protective antibodies [45]. We initiated experiments to determine the PEDV VLP immunogenicity in mice. A high anti-PEDV-neutralizing antibody level was induced in mice vaccinated with VLPs. Because no porcine model is available in our laboratory, we did not carry out further tests on pigs. This study was the foundation for further explorations that will test whether this vaccine is effective at preventing PEDV in pigs.
It is well known that IFN-γ is secreted by Th1 cells and regulates cellular immune responses and that IL-4 is secreted by Th2 cells and mediates the humoral immune response. The maturity of Th1 cells is also inhibited by IL-4. In our study, IL-4 levels were significantly enhanced relative to IFN-γ levels. However, IFN-γ levels were barely different from those of the control group. This demonstrated that Th1 cells were inhibited by the enhanced IL-4 levels and that the Th2 cell-mediated response was elicited by the PEDV VLPs. As mentioned above, IL-4 plays a leading role in the protection against PEDV infection. This conclusion is consistent with our previous observations [46]. The literature suggests that the format of antigen binding with the adjuvant should be considered and that different adjuvants should be assessed. Different antigen formats correspond to different antigen–antibody complex formats; consequently, the stimulated T cells should secrete different cytokines and elicit different types of immune responses. The Th2-type immune response primarily activates B cells, and specific anti-antigen antibodies are secreted. Consequently, the relevant pathogens are eliminated, and the host is protected. By contrast, a biased Th2-type immune response can cause immunopathology [47]. As such, this issue requires further study. Therefore, the ideal vaccine should be safe and effective for the host by containing the virus itself while minimizing deleterious effects in vaccinated animals.
In conclusion, this study developed PEDV VLPs assembled from viral S, M, and E proteins. The VLPs had superior immunogenicity and induced humoral immune responses in vaccinated mice. Additionally, higher levels of specific virus-neutralizing antibodies were obtained. We believe that VLP-based vaccines have the potential to prevent PEDV infections in the future.
Acknowledgements
This study was supported by the National Science and Technology Support program of China (No. 2015BAD12B04).
Author contributions
CW, FY, YG, SY, and XX designed the experiments. CW, FY, XZ, HW, HJ, CW, and NF performed the experiments. CW, FY, YZ, and TW analyzed the data. CW and FY wrote the manuscript. All authors reviewed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest, and all authors approved the final manuscript for publication.
Ethical approval
The mice experimentation protocol was approved by the Animal Welfare and Ethics Committee of the Veterinary Institute at the Academy of Military Medical Sciences under permit number SCXK-2014-0024. The environment and housing facilities satisfied the National Standards of Laboratory Animal Requirement (GB 14925-2011) of China.
Footnotes
Cuiling Wang and Feihu Yan have contributed equally to this work.
Contributor Information
Songtao Yang, Email: yst62041@163.com.
Xianzhu Xia, Email: xiaxzh@cae.cn.
References
- 1.Song D, Park B. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T, Takeyama N, Katsumata A, Tuchiya K, Kodama T, Kusanagi K. Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo LL, Masters PS. J. Virol. 2010;84:12872–12885. doi: 10.1128/JVI.01850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayanan K, Maeda A, Maeda J, Makino S. J. Virol. 2000;74:8127–8134. doi: 10.1128/JVI.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q. Emerg. Infect. Dis. 2012;8:1350–1353. doi: 10.3201/eid1803.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puranaveja S, Poolperm P, Lertwatcharasarakul P, Kesdaengsakonwut S, Boonsoongnern A, Urairong K, Kitikoon P, Choojai P, Kedkovid R, Teankum K, Thanawongnuwech R. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury B, Dastjerdi A, Doyle N, Frossard JP, Steinbach F. Virus Res. 2016;226:40–49. doi: 10.1016/j.virusres.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung HC, Lee JH, Nguyen VG, Huynh TM, Lee GE, Moon HJ, Park SJ, Kim HK, Park BK. Virus Res. 2016;226:14–19. doi: 10.1016/j.virusres.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JF, Sun DB, Wang CB, Shi HY, Cui XC, Liu SW, Qiu HJ, Feng L. Virus Genes. 2008;36:355–364. doi: 10.1007/s11262-007-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mole B. Nature. 2013;499:388–389. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- 12.Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. Can. Vet. J. 2015;56:149–152. [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 14.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kweon CH, Kwon BJ, Lee JG, Kwon GO, Kang YB. Vaccine. 1999;17:2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song DS, Oh JS, Kang BK, Yang JS, Moon HJ, Yoo HS, Jang YS, Park BK. Res. Vet. Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Eur. J. Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 18.Kang TJ, Kang KH, Kim JA, Kwon TH, Jang YS, Yang MS. Protein Expr. Purif. 2004;38:129–135. doi: 10.1016/j.pep.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Gerdts V, Zakhartchouk A. Vet. Microbiol. 2016 doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noad R, Roy P. Trends Microbiol. 2003;11:438–444. doi: 10.1016/S0966-842X(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Limas WA, Sekar K, Tyo KE. Curr. Opin. Biotechnol. 2013;24:1089–1093. doi: 10.1016/j.copbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Zheng X, Gai W, Zhao Y, Wang H, Wang H, Feng N, Chi H, Qiu B, Li N, Wang T, Gao Y, Yang S, Xia X. OncoTarget. 2016 [Google Scholar]
- 23.Sun DB, Feng L, Shi HY, Chen JF, Liu SW, Chen HY, Wang YF. Acta Virol. 2007;51:149–156. [PubMed] [Google Scholar]
- 24.Brian DA, Baric RS. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arevalo MT, Wong TM, Ross TM. J. Vis. Exp. 2016;112:e54041. doi: 10.3791/54041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makadiya N, Brownlie R, van den Hurk J, Berube N, Allan B, Gerdts V, Zakhartchouk A. Virol. J. 2016;13:57. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh J, Lee KW, Choi HW, Lee C. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jongwe TI, Chapman R, Douglass N, Chetty S, Chege G, Williamson AL. PLoS ONE. 2016;11:e0159141. doi: 10.1371/journal.pone.0159141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song D, Moon H, Kang B. Clin. Exp. Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frietze KM, Peabody DS, Chackerian B. Curr. Opin. Virol. 2016;18:44–49. doi: 10.1016/j.coviro.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 32.Delere Y, Wichmann O, Klug SJ, van der Sande M, Terhardt M, Zepp F, Harder T. Dtsch. Arztebl. Int. 2014;111:584–591. doi: 10.3238/arztebl.2014.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Y, Fan J, Huang W, Zhao C, Wang Y, Kong FT, Kong W, Jiang C. Biotechnol. Appl. Biochem. 2016;63:362–370. doi: 10.1002/bab.1379. [DOI] [PubMed] [Google Scholar]
- 34.Chroboczek J, Szurgot I, Szolajska E. Acta Biochim. Pol. 2014;61:531–539. [PubMed] [Google Scholar]
- 35.Lee YT, Kim KH, Ko EJ, Lee YN, Kim MC, Kwon YM, Tang Y, Cho MK, Lee YJ, Kang SM. Clin. Exp. Vaccine Res. 2014;3:12–28. doi: 10.7774/cevr.2014.3.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen HG, Halbur PG, Opriessnig T. J. Gen. Virol. 2012;93:1345–1355. doi: 10.1099/vir.0.039552-0. [DOI] [PubMed] [Google Scholar]
- 37.Glenn GM, Smith G, Fries L, Raghunandan R, Lu H, Zhou B, Thomas DN, Hickman SP, Kpamegan E, Boddapati S, Piedra PA. Vaccine. 2013;31:524–532. doi: 10.1016/j.vaccine.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Kim SH, Chen S, Jiang X, Green KY, Samal SK. J. Virol. 2014;88:9718–9727. doi: 10.1128/JVI.01570-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Chandramouli S, Medina-Selby A, Coit D, Schaefer M, Spencer T, Brito LA, Zhang P, Otten G, Mandl CW, Mason PW, Dormitzer PR, Settembre EC. Vaccine. 2013;31:3872–3878. doi: 10.1016/j.vaccine.2013.06.062. [DOI] [PubMed] [Google Scholar]
- 40.Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, Adegnika AA, Mordmuller B, Issifou S, Kremsner PG, Sacarlal J, Aide P, Lanaspa M, Aponte JJ, Machevo S, Acacio S, Bulo H, Sigauque B, Macete E, Alonso P, Abdulla S, Salim N, Minja R, Mpina M, Ahmed S, Ali AM, Mtoro AT, Hamad AS, Mutani P, Tanner M, Tinto H, D’Alessandro U, Sorgho H, Valea I, Bihoun B, Guiraud I, Kabore B, Sombie O, Guiguemde RT, Ouedraogo JB, Hamel MJ, Kariuki S, Oneko M, Odero C, Otieno K, Awino N, McMorrow M, Muturi-Kioi V, Laserson KF, Slutsker L, Otieno W, Otieno L, Otsyula N, Gondi S, Otieno A, Owira V, Oguk E, Odongo G, Woods JB, Ogutu B, Njuguna P, Chilengi R, Akoo P, Kerubo C, Maingi C, Lang T, Olotu A, Bejon P, Marsh K, Mwambingu G, Owusu-Agyei S, Asante KP, Osei-Kwakye K, Boahen O, Dosoo D, Asante I, Adjei G, Kwara E, Chandramohan D, Greenwood B, Lusingu J, Gesase S, Malabeja A, Abdul O, Mahende C, Liheluka E, Malle L, Lemnge M, Theander TG, Drakeley C, Ansong D, Agbenyega T, Adjei S, Boateng HO, Rettig T, Bawa J, Sylverken J, Sambian D, Sarfo A, Agyekum A, Martinson F, Hoffman I, Mvalo T, Kamthunzi P, Nkomo R, Tembo T, Tegha G, Tsidya M, Kilembe J, Chawinga C, Ballou WR, Cohen J, Guerra Y, Jongert E, Lapierre D, Leach A, Lievens M, Ofori-Anyinam O, Olivier A, Vekemans J, Carter T, Kaslow D, Leboulleux D, Loucq C, Radford A, Savarese B, Schellenberg D, Sillman M, Vansadia P, Engl N. J. Med. 2012;367:2284–2295. [Google Scholar]
- 41.Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Expert Rev. Vaccines. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 42.Kushnir N, Streatfield SJ, Yusibov V. Vaccine. 2012;31:58–83. doi: 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cruz DJ, Kim CJ, Shin HJ. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun DB, Feng L, Shi HY, Chen JF, Cui XC, Chen HY, Liu SW, Tong YE, Wang YF, Tong GZ. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Chen J, Shi H, Chen X, Shi D, Feng L, Yang B. Virol. J. 2012;9:225. doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suo S, Ren YD, Li GX, Zarlenga D, Bu RE, Su DD, Li XL, Li PC, Meng FD, Wang C, Ren XF. Virus Res. 2012;167:259–266. doi: 10.1016/j.virusres.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, Peters CJ, Couch RB. PLoS ONE. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]


