Abstract
Since late 2010, porcine epidemic diarrhea virus (PEDV) has been re-emerging in immunized swine herds with devastating impact in the Hebei province of China. Seven prevailing strains of PEDV were isolated from fecal samples out of piglets suffering from severe diarrhea. The M gene of the seven PEDV isolates encompasses an open reading frame of 681 nucleotides, encoding a protein of 226 amino acids. The seven PEDV isolates showed 99.4–99.9 % nucleotide sequence identity and 98.2–99.1 % deduced amino acid identity. When compared with other Chinese isolates and foreign isolates, the seven isolates showed high nucleotide identity with the Thailand isolate M-NIAH1005 (99.6–99.9 %) and Korea isolate PFF188 (99.7–100 %), but low identity with other Chinese isolates (96.6–99.1 %) and with the vaccine strain CV777 used in China (97.8–98.2 %). Phylogenetic analyses showed that all seven Chinese field isolates were grouped together in the same cluster. Although CV777 was also separated into the same cluster with the seven isolates, they were belonged to different sub-cluster. These results showed that the seven prevailing isolates in China are closely related phylogenetically to each other and have close relationships with the Korean strain PFF188 and Thailand strain M_NIAH1005. However, they differ genetically from other Chinese isolates and the vaccine strain CV777. Therefore, a more efficient vaccine strain should be chosen to prevent outbreaks of PEDV in China.
Keywords: Porcine epidemic diarrhea virus, M protein genes, Genotype, Phylogenetic relationship
Introduction
Porcine epidemic diarrhea virus (PEDV), a member of the family Coronaviridae, is an enveloped single-stranded RNA virus [7, 8, 17]. Its genome contains genes for the spike (S), membrane (M), small membrane (sM), open reading frame (ORF) 3, and nucleocapsid (N) proteins [2, 4]. The M protein is a structural membrane glycoprotein and the most abundant of all the envelope proteins. It has a short amino-terminal domain on the outside of the virion and a long carboxy-terminal domain on the inside. The M protein plays a central role in PEDV assembly. Together with the sM protein, it is responsible for assembly of the PEDV envelope [1, 9, 21]. In addition, the M protein directs the incorporation of the S protein [13, 15] and the nucleocapsid [12] into the budding particle.
PEDV causes severe entero-pathogenic diarrhea in piglets, especially in neonates, and the disease has a high mortality rate which can reach 80 % in certain situations [19]. PEDV was first reported in Belgium and the United Kingdom in 1978. Since then, outbreaks of the disease have been reported in several swine-raising countries, most notably in Europe and Asia, including Japan, China, and Korea [11]. It causes heavy economic losses in the European and Asian swine industry and is an important gastrointestinal disease [7, 16]. Several vaccines have been applied to the regions with a high prevalence of PEDV, but these vaccines have not been completely effective in preventing the disease [6, 18]. Since early 2006, PEDV has been re-emerging in immunized swine herds in China [5, 20]. In late 2010, outbreaks of PED occurred in HeBei province. Pigs of all ages were affected and losses from this disease outbreak were extensive. Most of the affected farms lost 100 % of their newborn piglets. In order to identify the genetic heterogeneity of the prevailing strains in China, PEDV strains were isolated from fecal samples from piglets infected with PEDV in 2010. The M genes were cloned and sequenced for analysis of genetic diversity, and the phylogenetic relationships of the field isolates with other PEDV reference strains were also analyzed.
Materials and methods
Virus and reagents
Eighty-two fecal samples were collected from piglets of 10 farms where the piglets presented clinical signs related to the PED definition and were from affected herds with severe diarrhea. Seven PEDVs were isolated from the feces that presented positive for PEDV. These isolates were used for nucleotide sequence analysis, amino acid sequence analysis, and phylogenetic analysis in this study, and are described in Table 1. Restriction endonucleases, polymerases, and ligases were purchased from TaKaRa Biotechology (Dalian, China) Co., Ltd.
Table 1.
Seven Chinese isolates and reference strains of PEDV used for sequence alignment, sequence analysis, and phylogenetic analysis
| Strain | Location and temporal information | Accession no. | Strain | Location and temporal information | Accession no. |
|---|---|---|---|---|---|
| 83P-5 | Japan, 2011 | AB618615 | LJB/03 | China, 2003 | AY608890 |
| 83P-5,100th | Japan, 2011 | AB618618 | LZC | China, 2007 | EF185992 |
| JMe2 | Japan, 1997 | D89752 | QH | China, 2005 | AY974335 |
| Chinju99 | Korea, 2006 | DQ845249 | YM/2007 | China, 2007 | EU302820 |
| KPED-9 | Korea, 1999 | AF015888 | HN/XYYYP | China, 2007 | EU287429 |
| V2501 | Korea, 2011 | FJ687458 | CH/SHH/06 | China, 2006 | EU033966 |
| PFF188 | Korea, 2011 | FJ687462 | CH/JSX/06 | China, 2006 | EU033967 |
| M2366 | Korea, 2011 | FJ687457 | CH/IMT/06 | China, 2006 | EU033965 |
| M1763 | Korea, 2011 | FJ687455 | CH/HNCH/06 | China, 2006 | EU033963 |
| M1595 | Korea, 2011 | FJ687452 | CH/HLJH/06 | China, 2006 | EU033964 |
| e1642 | Korea, 2011 | FJ687453 | JS/2004 | China, 2004 | AY653205 |
| CPF259 | Korea, 2011 | FJ687465 | BJ2010 | China, 2010 | JF690778 |
| BIF118 | Korea, 2011 | FJ687460 | HB/BD | China, 2011 | JF690777 |
| BI1108 | Korea, 2011 | FJ687451 | HB/FN | China, 2011 | JF508465 |
| BI981 | Korea, 2011 | FJ687450 | HB/HS | China, 2011 | JF690779 |
| Br1/87 | United Kingdom, 2006 | Z24733 | HB/GY | China, 2011 | JN400910 |
| M_NIAH1005 | Thailand, 2008 | EU542418 | HB/LF | China, 2011 | JN400909 |
| M_NIAH1795 | Thailand, 2008 | EU542415 | HB/QY | China, 2011 | JN400911 |
| M_NIAH380 | Thailand, 2008 | EU581712 | CV777 | Belgium,1998 | AF353511 |
Extraction of genomic PEDV RNA
Each fecal sample was centrifuged using a Beckman F3602 rotor at 2,000 g at 4 °C for 5 min. The supernatant was used for the extraction of the viral genomic RNA using Trizol reagent (Invitrogen, USA) based on the manufacturer’s instructions. The extracted RNA was resuspended in RNase-free water and was stored at −80 °C.
Primers for RT-PCR
Primers were designed on the basis of the published sequence of the M gene of PEDV. The forward and reverse primers designed to amplify the whole sequence of M gene of PEDV isolates were MP1 5′-GGATCCATGTCTAACGGT-3′ and MP2 5′-AAGCTTTCTGTTTAGACTAAAT-3′. The primers were synthesized commercially.
RT-PCR
The conditions for amplification included a reverse transcription process. Reverse transcription was carried out using the primer Oligo (dT) 18. The mixture of 1 μl virus RNA, 1 μl of 50 pM Oligo (dT)18, and 5 μl of RNase-free dH2O was incubated at 70 °C for 5 min and placed on ice for at least 2 min. Subsequently, 2 μl of 5× RT buffer, 0.5 μl of 10 mM dNTP, 0.25 μl of RNase inhibitor (40 U/μl), and 0.25 μl of M-MLV reverse transcriptase (200 U/μl) were added to initiate the reaction at 42 °C for 60 min. The reaction was stopped by heating at 70 °C for 15 min. The cDNA was either stored at −20 °C or amplified immediately.
The M genes of the seven field isolates were amplified by PCR using primers MP1 and MP2. The PCR conditions included an initial denaturation step at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 40 s, annealing at 46 °C for 40 s, extension at 72 °C for 60 s, then a final extension at 72 °C for 8 min. The PCR products were analyzed by electrophoresis in 1 % agarose gel containing ethidium bromide and visualized under UV light.
Cloning and sequencing
The M gene fragments obtained using RT-PCR were separated by gel electrophoresis and purified using a Gel Extraction Mini Kit (TIANGEN BIOTECH (Beijing) Co., Ltd.) according to the manufacturer’s recommendation. The purified product was cloned into a pMD18-T plasmid DNA vector, and the resulting plasmid was named pMD-M. The recombinant plasmid was used to transform competent Escherichia coli JM109. The positive clones were selected by their lacZ-negative phenotypes and verified by restriction enzyme digestion, PCR, and DNA sequencing.
Nucleotide sequence accession number
The complete nucleotide sequences of the M genes of the seven PEDV isolates have been deposited in the GenBank Database. Their accession numbers are given in Table 1.
Sequence analysis
Comparison and phylogenetic analysis of the nucleotide sequences of the M genes of the seven PEDV isolates with those of other PEDV strains were performed with PHYLIP v3.69 software. The reference strains used for the sequence analysis are described in Table 1. Phylogenetic relationships were determined by neighbor-joining method with PHYLIP v3.69 and reliability of topologies was estimated by performing bootstrap analysis with 1,000 replicates. TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to predict the transmembrane helices in M proteins. Antigenic index was evaluated by Jameson–Wolf method.
Results
Pathogen detection
The 82 fecal samples were submitted to pathogen detection and isolation. Detection results showed that 51 out of 82 (62.2 %) were positive for PEDV, 20 out of 82 (24.4 %) for transmissible gastroenteritis virus (TGEV), 14 out of 82 (17.1 %) for porcine rotavirus (PRV), and 38 out of 82 (46.3 %) for E. coli. E. coli positive cases were often positive for one other viral pathogen. From the 51 PEDV positive feces, Seven PEDV strains were isolated and their M genes were cloned.
M gene sequence analysis
Sequence analysis indicated that the nucleotide sequence of the M gene ORF of the seven PEDV isolates was 681 bp in length and encoded 226 amino acids. There were no deletions or insertions in the ORFs of the seven Chinese isolates. Compared with PEDV CV777, HB–HS had 12 nucleotide mutations, BJ2010 and HB-LF had 14 nucleotide mutations, and HB/FN, HB/BD, HB/GY, and HB/QY had 13 nucleotide mutations. These nucleotide mutations would lead to changes in the predicted amino acid sequences of the PEDV isolates. Compared with PEDV CV777, all seven field isolates had three amino acid changes (from E to Q at 13, from V to A at 42, from A to S at 214), in addition, BJ2010 had one further amino acid change (from T to A at 27), HB/FN had one amino acid change (from K to N at 48), HB/QY had one amino acid change (from Y to H at 198), HB/BD and HB/GY had one amino acid change (from S to A at 111), and HB/LF had two amino acid changes (from S to A at 111, from K to R at 202).
The M protein of the seven isolates had three transmembrane helices. They located in amino acid 20–38, 43–65, and 75–97 plots, respectively. 1–19 and 66–74 plots were outside the membrane while 39–42 and 98–226 plots were inside. The regions 2–13, 102–107, 112–118, 182–188, 200–206, and 216–221 had high antigenic index. Among the amino acid changes, those at 13 and 202 were located in the antigenic regions 2–14 and 200–206, respectively. It is uncertain now whether these changes led to the antigenic difference between the seven Chinese isolates and vaccine strain CV777. Further studies should focus on the relationship between them.
Sequence homology analysis
The nucleotide and deduced amino acid sequences of our isolates were compared with those of the reference strains. The sequence homology among these seven isolates was 99.4–99.9 % at the nucleotide level. When compared with the Chinese reference strains, the seven isolates had 96.6–99.1 % sequence homology with them: JS/2004 had 98.7–99.1 % sequence homology with the seven isolates, while LZC had 96.6–97.1 % homology with them. All seven isolates had low nucleotide identity (97.8–98.2 %) with CV777. HB/GY and HB/LF had only 97.8 % sequence homology with CV777. When the field isolates were compared with reference strains from other countries, the nucleotides identity values were 96.2–100 %. The highest nucleotides identity was observed between the seven field isolates and PFF188 and M-NIAH1005. The seven isolates had 99.7–100 % sequence homology with PFF188 and 99.6–99.9 % sequence homology with M-NIAH1005. HB–HS had 100 % sequence homology with PFF188 and 99.9 % with M-NIAH1005, while HB–GY and M-NIAH380 only showed a low nucleotide identity of 96.2 %. The deduced amino acid sequences of the field isolates showed 98.2–99.1 % homology, and they showed homology of 94.7–100 % with the deduced amino acid sequences of the reference strains.
Phylogenetic relationships
To analyze the phylogenetic relationships of the seven PEDV isolates with the reference strains isolated in various parts of the world, we constructed a neighbor-joining phylogenetic tree using the M gene sequences. A representative tree for the M gene is shown in Fig. 1. The seven PEDV isolates and reference strains were separated into three potential clusters. All seven field isolates were segregated into the same cluster. In terms of the topology of the phylogenetic tree, the seven PEDV isolates might have a common origin.
Fig. 1.
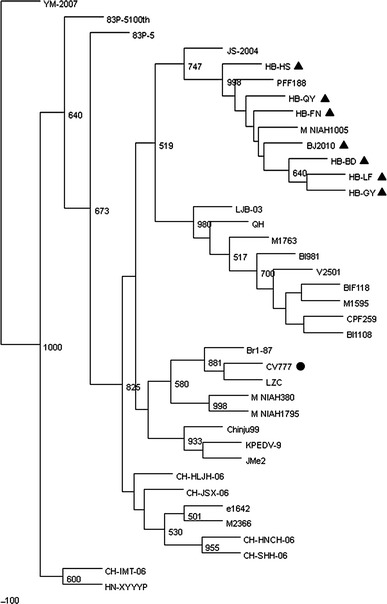
Phylogenetic tree of the nucleotide sequences of PEDV isolates based on the M gene. Sequences of reference strains were obtained from the GenBank database. Genbank accession numbers are shown in Table 1. Tree topology was constructed using F84 model and bootstrap re-sampling (1,000 data sets) of the multiple alignments was used to test the statistical robustness of the trees obtained by NJ (using the program Neighbor from PHYLIP v3.69 package) ● vaccine strain ▲ epidemic strain
Discussion
Porcine epidemic diarrhea virus (PEDV) is a member of the coronavirus group causing watery diarrhea, dehydration, and high mortality in suckling pigs. Although the bi-combined attenuated vaccine against TGEV and PEDV infection is authorized for use on swine farms, PED still occurs in the immunized swine herds in China. Since late 2010, outbreaks of PED have occurred in Hebei province. Most of the affected farms lost 100 % of their newborn piglets. PEDV is known to have only one serotype, but the genome shows wide genetic diversity [3, 10, 22]. The structural M protein of PEDV plays an important role in the assembly process and induces virus neutralization antibodies [14]. Therefore it is necessary to investigate the prevalence of PEDV on the basis of phylogenetic analysis of the M gene of the field strains of PEDV in China.
This study was conducted to characterize PEDVs isolated from the fecal samples of piglets suffering from severe diarrhea in Hebei province of the People’s Republic of China. The initial aim of this study was to isolate and investigate the genetic diversity among field strains of PEDV in China. A large amount of sequence data on PEDVs isolated throughout the world has been published over the years and is now available for sequence comparison and phylogenetic analysis. In this study, seven Chinese field strains of PEDV were isolated. The complete nucleotide and deduced amino acid sequences of the M gene were determined and compared with those of the reference strains. None of the seven isolates were found to have sequence insertions or deletions in the M genes, although all the PEDV isolates investigated in this study had nucleotide mutations. Analysis of sequence homology showed that the seven PEDV isolates were closely related to PFF188, M-NIAH1005, and JS-2004. Furthermore, HB–HS and PFF188 had identical nucleotide sequences, although they were isolated in different countries at different times. This suggests that HB–HS might have been generated naturally from PFF188. However, lower amino acid identity was observed between the seven PEDV isolates and the Chinese isolates CH-IMT-06, HN/XYYYP, YM/2007. Phylogenetically, they belonged to three different clusters. Although the seven isolates and the other Chinese isolates belonged to the same cluster, they were segregated into different sub-clusters. These results showed that a new genotype of PEDV prevails in China now. None of the seven PEDV isolates was phylogenetically close to the vaccine strain CV777 that is used in China. In fact, they were segregated into different sub-clusters. In addition, the seven Chinese isolates also had a change in the antigenic regions 2–14 though there has no scientific data to prove whether this change had brought about antigenic variation. All these results show that, in spite of the regular use of live vaccines in pigs, the seven field isolates of PEDV were of a different genotype able to cause outbreaks in immunized pigs in Hebei province. In conclusion, genetic variation exists in the M gene of the prevalent Chinese PEDV field isolates, and a more appropriate vaccine strain should be chosen to prevent PEDV outbreaks in China.
Acknowledgments
This study was supported by the research foundation of the Education Bureau of Hebei Province (No. 2011180), China.
References
- 1.Baudoux P, Besnardeau L, Carrat C, Rottier P, Charley B, Laude H. Interferon alpha inducing property of coronavirus particles and pseudoparticles. Adv. Exp. Med. Biol. 1998;440:377–386. doi: 10.1007/978-1-4615-5331-1_49. [DOI] [PubMed] [Google Scholar]
- 2.Bridgen A, Duarte M, Tobler K, Laude H, Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 1993;74:1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- 3.Bridgen A, Kocherhans R, Tobler K, Carvajal A, Ackermann M. Further analysis of the genome of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1998;440:781–786. doi: 10.1007/978-1-4615-5331-1_101. [DOI] [PubMed] [Google Scholar]
- 4.Chang SH, Bae JL, Kang TJ, Kim J, Chung GH, Lim CW, Laude H, Yang MS, Jang YS. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14(2):295–299. [PubMed] [Google Scholar]
- 5.Chen JF, Sun DB, Wang CB, Shi HY, Cui XC, Liu SW, Qin HJ, Feng L. Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes. 2008;36(2):355–364. doi: 10.1007/s11262-007-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi HJ, Kim JH, Lee CH, Ahn YJ, Song JH, Baek SH, Kwon DH. Antiviral activity of quercetin 7-rhamnoside against porcine epidemic diarrhea virus. Antiviral Res. 2009;81:77–81. doi: 10.1016/j.antiviral.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debouck P, Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980;41:219–223. [PubMed] [Google Scholar]
- 8.Ducatelle R, Coussement W, Pensaert M, Debouck P, Hoorens J. In vivo morphogenesis of a new porcine enteric coronavirus, CV777. Arch. Virol. 1981;68:35–44. doi: 10.1007/BF01315165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godeke GJ, de Haan CA, Rossen JW, Vennema H, Rottier PJ. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74(3):1566–1571. doi: 10.1128/JVI.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubota S, Sasaki O, Amimoto K, Okada N, Kitazima T, Yasuhara H. Detection of porcine epidemic diarrhea virus using polymerase chain reaction and comparison of the nucleocapsid protein genes among strains of the virus. J. Vet. Med. Sci. 1999;61(7):827–830. doi: 10.1292/jvms.61.827. [DOI] [PubMed] [Google Scholar]
- 11.Kweon CH, Kwon BJ, Jung TS, Kee YJ, Hur DH, Hwang EK, Rhee JC, An SH. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J. Vet. Res. 1993;33:249–254. [Google Scholar]
- 12.Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74(17):8127–8134. doi: 10.1128/JVI.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen VP, Hogue BG. Protein interactions during coronavirus assembly. J. Virol. 1997;71(12):9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni Yan-xiu, Lin Ji-huang, He Kong-wang, Huan Hong-hua. Research overview of porcine epidemic diarrhea virus. Anim. Husb. Vet. Med. 2001;33(1):38–40. [Google Scholar]
- 15.Opstelten DJ, Raamsman MJ, Wolfs K, Horzinek MC, Rottier PJ. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131(2):339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pensaert MB. Porcine epidemic diarrhea virus. In: Dunn HW, editor. Disease of Swine. 8. Ames: Iowa State University Press; 1999. pp. 179–185. [Google Scholar]
- 17.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song HJ, Chae SW, Yoon KA, Park JS, Choi HJ. Antiviral activity of Zanthoxylum species against porcine epidemic diarrhea virus. J. Cosmetic. Public Health. 2010;6(2):42–44. [Google Scholar]
- 19.Song SS, Kang BK, Lee SS, Yang JS, Moon HJ, Oh JS, Ha GW, Jang YS, Park BY. Use of an internal control in a quantitative RT-PCR assay for quantitation of porcine epidemic diarrhea virus shedding in pigs. J. Virol. Methods. 2006;133(1):27–33. doi: 10.1016/j.jviromet.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Sun Dong-bo, Feng Li, Shi Hong-yan, Chen Jian-fei, Cui Xiao-chen, Tong You-en. Preparation and characterization of the monoclonal antibodies against neutralizing epitope region of PEDV s protein. Chinese J. Prev. Vet. Med. 2007;29(11):887–890. [Google Scholar]
- 21.Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo SG, Hernandez M, Krell PJ, Nagy EE. Cloning and sequence analysis of the spike gene of porcine epidemic diarrhea virus Chinju99. Virus Genes. 2003;26(3):239–246. doi: 10.1023/A:1024443112717. [DOI] [PMC free article] [PubMed] [Google Scholar]


