Abstract
Infectious bronchitis virus (IBV) affects both vaccinated and unvaccinated flocks worldwide, with a significant impact on the poultry industry. The aim of the present study is to characterize an emerging variant pathogenic IBV originating from field outbreaks in vaccinated Egyptian layer flock. Samples were collected from disease-suspected flock with a history of administration of live and inactivated IBV vaccines (Ma5 type). Virus propagation in embryonated chicken eggs (ECEs), after three successive passages, revealed typical IBV lesions such as curling and dwarfism. The reported isolate was identified by a real-time reverse transcriptase PCR assay targeting nucleocapsid (N) gene and, further characterized by full-length spike (S1) gene sequencing. Phylogenetic analysis revealed clustering of the isolated virus within 4/91 genotype of GI-13 lineage. Deduced amino acid sequences identity revealed 75–76% and 88–90% similarity with the currently used classic (H120, Ma5, and M41) and variant vaccine strains (4/91 and CR88) in Egypt, respectively. Recombination analysis gave an evidence for distinct patterns of origin for the studied isolate providing another example of intra-genotypic recombination among IBVs and the first example of recombination within the GI-13 lineage in the Egyptian field. The studied isolate (IBV/CK/EG/Fadllah-10/2019) emerged as a result of recombination between the variant group (Egy/var I genotype, GI-23 lineage) as a major parent and the CR88 variant vaccine strain (4/91 genotype, GI-13 lineage) as minor parent. Our data suggest that both mutation and recombination may be contributing to the emergence of IBV variants which ascertain the importance of disease monitoring in vaccinated flocks as well as re-appropriation for the current vaccine strategies.
Electronic supplementary material
The online version of this article (10.1007/s11262-019-01693-9) contains supplementary material, which is available to authorized users.
Keywords: Infectious bronchitis virus, Curling and dwarfism, Identity, Recombination, Emergence, Monitoring
Infectious bronchitis (IB) is a highly contagious disease of poultry, caused by infectious bronchitis virus (IBV) which replicates in a wide variety of epithelial cells of respiratory, renal, reproductive, and digestive tissues [1]. IB is considered the most important viral disease in countries which are free from Avian Influenza and/or Newcastle Disease [2]. IBV belongs to genus Gammacoronavirus within the Coronaviridae family [3], has positive-sense single-stranded RNA genome (27.6 kb) which encodes four major structural proteins; spike glycoprotein (S), membrane glycoprotein (M), envelope protein (E), and phosphorylated nucleocapsid protein (N) [4]. The S1 subunit of spike (S) glycoprotein gene contains epitopes for virus neutralization, cell attachment, and serotype specificity [5–7].
Genetic classification and evolutionary analysis of IBV variants are mainly based on S1 gene sequencing [8, 9]. The latest classification for IBV recognized six main genotypes (GI–GVI), 32 viral lineages (1–32), and a number of inter-lineage recombinants worldwide [10]. IBV was first reported in Egypt in 1954 [11]. By then, several reports have confirmed the disease circulation with continuous genetic diversity and evolution [12–26]. Despite mass vaccination strategies in Egypt utilizing Mass-type (H120, M41, and Ma5) and 793B-type(CR88 and 4/91) of vaccine, variant IBVs still devastate the poultry industry periodically. During the years 2010 to 2018, majority of the circulating IBV strains in the Egyptian poultry industry were clustered within GI-23 lineage that were further sub-categorized into Egy/variant I and Egy/variant II of IS/1494 and IS/885 origin, respectively [10, 23, 24]. Indeed, the IBVs pose a significant economic impact on poultry industry and, therefore, disease consequences are devastating the national economy of Egypt. The aim of the present study was to (1) characterize newly emerged variant IBV or escape mutant from a breeder layer flock and (2) determine the genetic divergence between this circulating field variant and the currently used classic and variant vaccine strains in Egypt.
Despite a history of vaccination using live attenuated and inactivated Ma5 vaccine (Table 1), clinical infectious bronchitis disease was suspected in a 85-week-old layer flock (White Bovans breeds) exhibiting respiratory signs along with high mortality and morbidity. The affected flock was raised in cages at Ismailia governorate, Egypt. Respiratory tract tissues (trachea) collected on 15th January 2019, showed congestion with caseous exudates, cloudiness, and turbidity of air sacs. Furthermore, degeneration of the ovary and swollen oviducts associated with egg peritonitis was observed. For virus isolation and propagation, a volume of 0.2 ml of tracheal homogenate was inoculated into the allantoic cavity of specific pathogen-free embryonated chicken eggs (SPF ECEs) (9- to 11-day old) [27], kindly obtained from Kom Oshim farm, Fayoum, Egypt. Inoculated eggs were monitored daily by candling for embryonic mortality. Two days after inoculation, the allantoic fluid was harvested and further passaged into another two successive passages.
Table 1.
Vaccination regime for the affected flock
| Day | Vaccine | Route of vaccination |
|---|---|---|
| 1st day | VAXXITEK® HVT + IBDV | S/C injection |
| 7th day | NDV clone 30 + IBV Ma5 (live attenuated) | Eye drop |
| 9th day | Inactivated H9N2 AIV | S/C injection |
| 15th day | Inactivated H5N1 AIV | S/C injection |
| 18th day | IBDV D78 | Eye drop |
| 28th day | NDV clone 30 + IBV Ma5 (live attenuated) | Eye drop |
| 35th day | Modified live ILTV | Eye drop |
| 40th day | Live Pox vaccine | Wing web |
| 49th day | NDV clone 30 + IBV Ma5 (live attenuated) | Eye drop |
| 60th day | Inactivated H9N2 AIV | S/C injection |
| 65th day | Inactivated H5N1 AIV | S/C injection |
| 70th day | NDV clone 30 + IBV Ma5 (live attenuated) | Eye drop |
| 91st day | NDV clone 30 + IBV Ma5 (live attenuated) | Eye drop |
| 115th day | Inactivated IBV, NDV and EDS | I/M injection |
Viral RNA was extracted from the allantoic fluid of 3rd passage using a QIAamp viral RNA mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Detection of IBV was conducted using a real-time reverse transcriptase PCR based on the highly conserved N gene, as described previously [28]. Amplification of full-length spike (S1) gene was conducted as described previously [25] using the Mx3005Pe system (Stratagene, Santa Clara, CA). PCR products were analyzed by agarose gel electrophoresis (1%) and then purified using a QIAquick Gel Extraction Kit (Qiagen) following the manufacturer’s instructions. Sequencing was carried out using BigDye Terminator v3.1 sequencing kit (Applied Biosystems) and an automated sequencer (ABI, 3130, Applied Biosystems, Foster City, CA). The quality of obtained sequence (S1 gene) was checked, assembled, edited using BioEdit software version 7.0.4.1 [29], and submitted to GenBank using BankIt tool of the GenBank (http://www.ncbi.nlm.nih.gov/WebSub/?tool=genbank), and obtained the accession number KP729419.
Deduced amino acid sequences alignment for the S1 subunit of the spike protein was performed by comparing with commonly utilized commercial vaccine strains in Egypt (Ma5, Ma41, H120, CR88, and 4/91). Phylogenetic analysis was conducted to explain the phylogenetic relationships with high-level of clustering pattern dependent on the full-length S1 gene between the reported isolate and recently described strains from the Middle East including Egypt and other parts of the world.
For maximum-likelihood analysis of the phylogenetic relationship, Kimura 2-parameter model of nucleotide substitution was used with gamma rate distribution, which was the best-fitting substitution model overall for the three clades, as determined by the substitution model-fitting function in MEGA 7 [30]. Trees were finally visualized and annotated using FigTree v1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree/). For recognition of recombination occasions, sequence alignments were conducted, in barrel with different available viruses resembling genotypes GI-1, GI-13, GI-19, and GI-23. Recombination Detection Program 4 (RDP4) software suite [31] were used to identify recombination events in the full-length S1 gene of IBV/CK/EG/Fadllah-10/2019 isolate through detection of breaking points using specific algorithms implemented in RDP4; RDP, Genecov, Bootscan, Maxchi, Chimaera, Siscan, and 3Seq with the highest acceptable P value adjusted to 0.05.
Here, we present the isolation and integrative genetic analysis that maps the emergence of 4/91 (793B) genotype in Egypt during 2019. Virus propagation in embryonated chicken eggs (ECEs), after three successive passages, revealed typical IBV lesions such as curling and dwarfism. After identification of IBV using a N gene-targeted real-time RT-PCR assay (Ct 12.22), the isolated IBV strain (IBV/CK/EG/Fadllah-10/2019) was genotyped based on sequencing and subsequent sequence analysis of the full-length S1 gene. The obtained nucleotide (1617bp) and corresponding amino acid sequences were aligned and compared with reference and vaccine strains (H120, Ma5, M41, 4/91, and CR88) including previously described IBV variants in Egypt and neighboring countries.
Compared to currently used vaccines in Egypt, the characterized virus shared different levels of nucleotide and amino acid sequence identities which were between 75 and 90% (Tables S1 and S2). The S1 protein contained hypervariable regions (HVRs) associated with serotype specificity and virus-neutralizing epitopes and located within the amino acid residues 38–67, 91–141, and 274–387 [32, 33]. A significant clustering of substitutions were detected in IBV/CK/EG/Fadllah-10/2019 in the three HVRs as compared to 4/91 and other vaccine strains. The prominent amino acid substitutions within the three HVRs in comparison with 4/91 vaccine were as follows at HVR1; D60F, V62A, S63G, D64Q, T69S, F70I, Y71H, E72W, Y74K, I76F, A79S. While at HVR2, the mutations were F115Y, S117N, Q118G, N131D, I135R, R139M, S141Y, F143I, and at HVR3 was only single-mutation (A281P) (Table 2). It is well known that even a small change in the amino acid sequence of the spike protein can result in generation of novel genotypes and/or serotypes that differ antigenically from the existing classic and variant vaccine strains [24, 25, 34]. In addition, it has been shown that changes as little as 5% in the S1 gene are able to alter the protective ability of a vaccine [33]. Moreover, residues N38, H43, P63, and T69 have been described to be critical for binding of the IBV spike protein to the chicken respiratory tract [35]. In the analyzed sequence of the isolate under study, residue substitutions associated with virus tropism were identified at positions N38P and G63Q in the receptor-binding site of the variant strains.
Table 2.
Sequence alignment of HVRs amino acid sequences of the reported isolate in this study compared to other reference strains/genotypes
| Strain | HVR1 (60–88) | HVR2 (115–140) | HVR3 (275–292) |
|---|---|---|---|
| 4/91 | VSVSDCTAGTFYESYNISAASVAMTVPPA | FKSQQGSCPLTGMIPQNHSIIVSARSGF | TNVSNASPNSGGVDTFQLY |
| CR88 | G. A……….. R…. S……. HN | .. N. L…………. IR. SA,. D. V | …………….. . |
| H120 | G. S. G.. V. IIHGGRVVN. S. I… A. SS | Y. H - -. G.. I… LQ. H.. R… MKN. Q | H. ETG. N.. PS.. QNI. T . |
| Ma5 | G. S. G.. V. IIHGGRVVN. S. I… A. SS | Y. H - -. G.. I… LQ. H.. R… MKN. Q | H. ETG. N.. PS.. QNI. T . |
| Ma41 | G. S. G.. V. IIHGGRVVN. S. I… A. SS | Y. H - -. G.. I… LQ. H.. R… MKN. Q | H. ETG. N.. PS.. QNI. T . |
| Fadllah-10/2019 | G. AGQ…. SIHW. K. F.. S….. A. YT | Y. NG………… D… R… M. Y. I | …… P……….. . |
A dot (.) indicates an identical amino acid. A dash (-) indicates an amino acid deletion
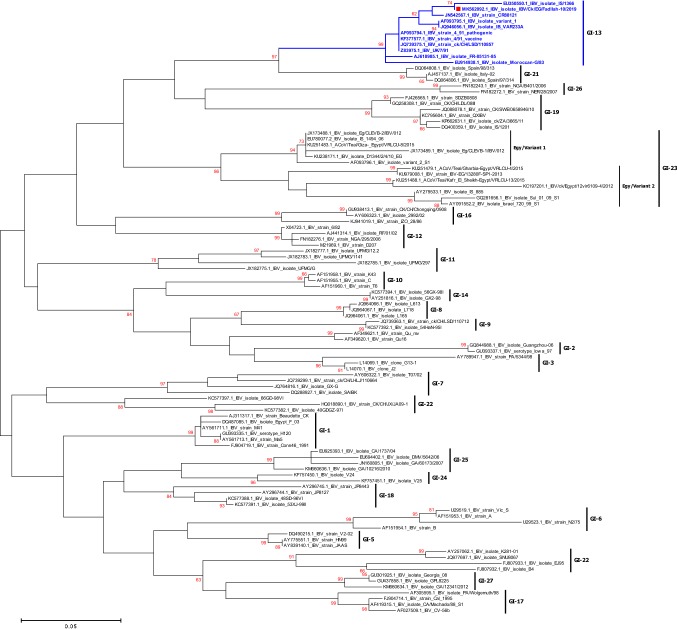
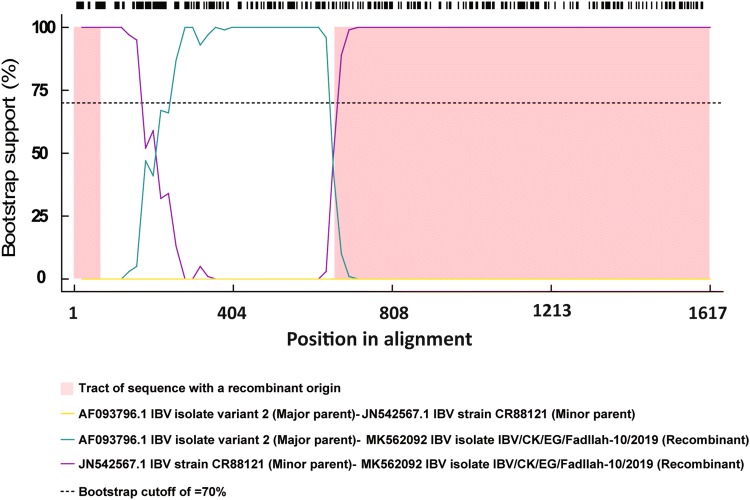
Previously Massachusetts-type strains were used for IBV vaccination programs in Egypt. Recently, the use of other strain types (e.g., 793/B serotype) has been officially authorized. However, the antigenic relatedness between GI-1, GI-16, and GI-23 lineages has not yet been analyzed, even though it may be useful to understand IBV epidemiology and to design specific control programs. Phylogenomic analysis based on S1 gene of the IBVs collected from Egypt and rest of the world indicated that Egyptian virus reported here was clustered within 4/91 genotype of GI-13 lineage (Fig. 1). Unique mechanism of RNA synthesis involving polymerase jumping and discontinuous transcription might be responsible for recombination in coronaviruses [36]. Studies reported that occurrence of recombination among different IBV field strains results in the emergence of new variants [37–39]. Many recombination events have been reported in different IBV strains, not only between field (wild type) and vaccine viruses but also among field viruses either within the same genotype (intra-genotypic) or between different genotypes (inter-genotypic) [40, 41], giving rise to new IBV genotypes [42]. Different available viruses resembling genotype GI-1, GI-12, GI-13, and the two subgroups of GI-23 were used in the analysis. The employed recombination detection methods revealed that IBV/CK/EG/Fadllah-10/2019 strain has undergone inter-genotypic recombination and different breakpoints within the S1 gene which might be a precursor for further GI-13 lineage evolution in Egypt in the near future. So, future studies are required to determine the pathobiological and clinical features of this virus in a chicken model. Also, the results showed that the reported isolate IBV/CK/EG/Fadllah-10/2019 is a recombinant virus which probably emerged from at least two different genotypes, including the Egy/Variant I genotype (GI-23 lineage) as a major parent and the CR88 vaccine strain (4/91 genotype of GI-13 lineage) as minor parents (Fig. 2).
Fig. 1.
Phylogenetic tree based on a full-length sequence of the S1 gene, showing the relationship between the circulating IBV Egyptian and worldwide genotypes relation to variant pathogenic KP729419 IBV/CK/EG/Fadllah-10/2019 isolate reported in this study. The robustness of individual nodes of the tree was assessed using 1000 replications of bootstrap re-sampling of the originally aligned nucleotide sequences. Scale bar represents the number of substitutions per site. The year of isolation and geographical origin of the virus sequences are included in the tree. Tree was constructed using maximum-likelihood method. The reported isolates in this study are marked with red square
Fig. 2.
Recombination detection analysis displaying possible recombination events predicted to have occurred in the S1 segment of the KP729419 IBV/CK/EG/Fadllah-10/2019 isolate
To recapitulate, the current study revealed the isolation and molecular identification of pathogenic 4/91 IBV collected from Egyptian vaccinated layer flock during 2019. Amino acids alignment revealed distinct mutations within HVRs compared to the commercially used vaccines. Likewise, phylogeny revealed that clustering of IBV/CK/EG/Fadllah-10/2019 strain within GI-13 lineage. Further analysis based on recombination detection analysis revealed the isolated strain undergoes distinct spots of recombination. Thus, the identified variant pathogenic 4/91 IBV may express as a new genotype that requires re-evaluation of vaccination strategies employed in Egypt. In conclusion, owing to high mutation rate and subsequent residue substations, there is an ongoing evolution among the circulating IBV variants in Egypt that results in frequent vaccination failure. A continuous disease monitoring and surveillance is required not only to elucidate sequence characteristics of prevailing strains but also to revise or appropriate vaccine strains and strategies accordingly.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all co-workers and colleagues in the Department of Virology, Faculty of Veterinary Medicine, Cairo University, Egypt, for their technical support.
Author contributions
MAR, RFE, AIB, and OKZ: Conceived the study. MAR, RFE, MMZ, SEL, MA, SAN, and MM: Performed research. MAR, RFE, AMG, SAN, and OKZ: Analyzed data. MAR, RFE, MZS, OKZ, and MM: Wrote the paper.
Funding
This work was financed by the Science and Technology Development Fund (STDF-STF, Project No. 24231), and British Council (172710323 and 332228521). The funding sources had no role in the study design, collection, or analysis of the data, writing of the manuscript, or in the decision to submit the manuscript for publication
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human and animal participants
International, national, and/or institutional guidelines for the care and use of poultry flocks were followed. The experiments were conducted with the approval of the Local Ethics Committee on Animal Experimentation in Faculty of Veterinary Medicine, Cairo University, Egypt.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 2.OIE . Manual of diagnostic tests and vaccines for terrestrial animals. Paris: OIE; 2004. Avian infectious bronchitis. [Google Scholar]
- 3.de Groot . Family Coronaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy, 9th report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press; 2012. pp. 806–828. [Google Scholar]
- 4.Stern DF, Sefton BM. Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J Virol. 1982;44(3):794–803. doi: 10.1051/vetres:2006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mockett AP, Cavanagh D, Brown TD. Monoclonal antibodies to the S1 spike and membrane proteins of avian infectious bronchitis coronavirus strain Massachusetts M41. J Gen Virol. 1984;65(12):2281–2286. doi: 10.1099/0022-1317-65-12-2281. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D, Davis PJ. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986;67(7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh D, Davis PJ, Cook JK, et al. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21(1):33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- 8.Lee CW, Hilt DA, Jackwood MW. Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene. J Vet Diagn Invest. 2003;15:344–348. doi: 10.1177/104063870301500407. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Junker D, Hock L, Ebiary E, Collisson EW. Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus. Virus Res. 1994;34:327–338. doi: 10.1016/0168-1702(94)90132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valastro V, Holmes EC, Britton P, et al. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed HN (1954) Incidence and treatment of some infectious viral respiratory diseases of poultry in Egypt. DVM Thesis. Cairo University, Cairo, Egypt
- 12.Sheble A, Sabry MZ, Davelaar FG, et al. Present status of infectious bronchitis in Egypt. J Egypt Vet Med Assoc. 1986;46:393–411. [Google Scholar]
- 13.Abdel-Moneim AS, Madbouly HM, Gelb JR, et al. Isolation and identification of Egypt/Beni-Seuf/01 a novel genotype of infectious bronchitis virus. Vet Med J Giza. 2002;50:1065–1078. [Google Scholar]
- 14.Abdel-Moneim AS, El-Kady MF, Ladman BS, et al. S1 gene sequence analysis of a nephropathogenic strain of avian infectious bronchitis virus in Egypt. Virol J. 2006;3:78. doi: 10.1186/1743-422X-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Susan S, El-Hady MM, Soliman YA. Isolation and characterization of nephropathogenic strain of infectious bronchitis virus in Egypt. J Am Sci. 2010;6:669–674. [Google Scholar]
- 16.Abdel-Moneim AS, Afifi MA, El-Kady MF. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch Virol. 2012;157:2453–2457. doi: 10.1007/s00705-012-1445-1. [DOI] [PubMed] [Google Scholar]
- 17.Selim K, Arafa AS, Hussein HA, et al. Molecular characterization of infectious bronchitis viruses isolated from broiler and layer chicken farms in Egypt during 2012. Int J Vet Sci Med. 2012;1:102–108. doi: 10.1016/j.ijvsm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussein AH, Emara MM, Rohaim MA, et al. Sequence analysis of infectious bronchitis virus IS/1494 like strain isolated from broiler chicken co-infected with Newcastle disease virus in Egypt during 2012. Int J Poult Sci. 2014;13:530–536. doi: 10.3923/ijps.2014.530.536. [DOI] [Google Scholar]
- 19.Samir M, Selim A, Arafa A, et al. Molecular diversity between field isolates and vaccinal strains of avian infectious bronchitis virus in Egypt. Glob Vet. 2014;13:820–827. [Google Scholar]
- 20.Abd El Rahman S, Hoffmann M, Lueschow D, et al. Isolation and characterization of new variant strains of infectious bronchitis virus in Northern Egypt. Adv Anim Vet Sci. 2015;3:362–371. doi: 10.14737/journal.aavs/2015/3.7.362.371. [DOI] [Google Scholar]
- 21.Sultan H, Abdel-Razik AG, Shehata AA, et al. Characterization of infectious bronchitis viruses circulating in Egyptian chickens during 2012 and 2013. J Vet Sci Med Diagn. 2015;4:5. [Google Scholar]
- 22.Awad EM, Arafa AS, El-Deeb AH, El-Sanousi AA. Molecular studies on infectious bronchitis virus isolated from broiler chickens in Damietta Governorate, Egypt. Zag Vet J. 2016;44:119–127. doi: 10.21608/zvjz.2016.7854. [DOI] [Google Scholar]
- 23.Zanaty A, Arafa AS, Hagag N, et al. Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J Virol. 2016;5:125–134. doi: 10.5501/wjv.v5.i3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanaty A, Naguib MA, El-Husseiny MH, et al. The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra-genotypic recombination. Arch Virol. 2016;161:3583–3587. doi: 10.1007/s00705-016-3042-1. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Sabour MA, Al-Ebshahy EM, Khaliel SA, et al. Isolation and molecular characterization of novel infectious bronchitis virus variants from vaccinated broiler flocks in Egypt. Avian Dis. 2017;61:307–310. doi: 10.1637/11566-121516-RegR. [DOI] [PubMed] [Google Scholar]
- 26.Rohaim MA, El Naggar RF, Helal AM, et al. Genetic diversity and phylodynamics of Avian coronaviruses in Egyptian wild birds. Viruses. 2019;11(1):57. doi: 10.3390/v11010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Office International des Epizooties . Manual diagnostic tests vaccines terrestrial animals (mammals, birds bees) Paris: Office International des Epizooties; 2018. [Google Scholar]
- 28.Meir R, Maharat O, Farnushi Y, Simanov L. Development of a real-time TaqMant RT-PCR assay for the detection of infectious bronchitis virus in chickens, and comparison of RT-PCR and virus isolation. J Virol Methods. 2010;163:190–194. doi: 10.1016/j.jviromet.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 30.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin DP, Murrell B, Golden M, et al. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1(1):vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore KM, Jackwood MW, Hilt DA. Identification of amino acids involved in a serotype and neutralization specific epitope within the S1 subunit of avian infectious bronchitis virus. Arch Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abozeid HH, Paldurai A, Khattar SK, et al. Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: evidence for genetic drift and genetic recombination in the circulating viruses. Infect Genet Evol. 2017;53:7–14. doi: 10.1016/j.meegid.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 35.De Wit JJ. Detection of infectious bronchitis virus. Avian Pathol. 2000;29:71–93. doi: 10.1080/03079450094108. [DOI] [PubMed] [Google Scholar]
- 36.Lai MM. Recombination in large rna viruses: coronaviruses. Semin Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ammayappan A, Upadhyay C, Gelb J, Jr, et al. Complete genomic sequence analysis of infectious bronchitis virus ark dpi strain and its evolution by recombination. Virol J. 2008;5:157. doi: 10.1186/1743-422X-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abro SH, Renstrom LH, Ullman K, et al. Characterization and analysis of the full-length genome of a strain of the european qx-like genotype of infectious bronchitis virus. Arch Virol. 2012;157:1211–1215. doi: 10.1007/s00705-012-1284-0. [DOI] [PubMed] [Google Scholar]
- 39.Abolnik C. Genomic and single nucleotide polymorphism analysis of infectious bronchitis coronavirus. Infect Genet Evol. 2015;32:416–424. doi: 10.1016/j.meegid.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Wang HN, Wang T, Fan WQ, Zhang AY, Wei K, Tian GB, Yang X. Complete genome sequence and recombination analysis of infectious bronchitis virus attenuated vaccine strain H120. Virus Genes. 2010;41(377–388):25. doi: 10.1007/s11262-010-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Z, Zhang T, Xu Q, Gao M, Chen Y, Wang Q, Zhao Y, Shao Y, Li H, Kong X, Liu S. Altered pathogenicity of a tl/CH/LDT3/03 genotype infectious bronchitis coronavirus due to natural recombination in the 5’-17 kb region of the genome. Virus Res. 2016;213:140–148. doi: 10.1016/j.virusres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackwood MW, Boynton TO, Hilt DA, McKinley ET, Kissinger JC, Paterson AH, Robertson J, Lemke C, McCall AW, Williams SM, Jackwood JW, Byrd LA. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.