Abstract
Porcine epidemic diarrhea virus (PEDV) is a member of the coronaviridae family, which can cause acute and highly contagious enteric disease of swine characterized by severe entero-pathogenic diarrhea in piglets. Currently, the vaccines of PEDV are only partially effective and there is no specific drug available for treatment of PEDV infection. To exploit the possibility of using RNA interference (RNAi) as a strategy against PEDV infection, five shRNA-expressing plasmids targeting the N, M, and S genes of PEDV were constructed and transfected into Vero cells. The cytopathic effect and MTS assays demonstrated that two shRNAs (pSilencer4.1-M1 and pSilencer4.1-N) were capable of protecting cells against PEDV invasion with very high specificity and efficiency. The two shRNA expression plasmids were also able to inhibit the PEDV replication significantly, as shown by detection of virus titers (TCID50/mL). A real-time quantitative RT-PCR further confirmed that the amounts of viral RNAs in cell cultures pre-transfected with these two plasmids were reduced by 95.0 %. Our results suggest that RNAi might be a promising new strategy against PEDV infection.
Keywords: Porcine epidemic diarrhea virus, RNA interference, Short hairpin RNA
Introduction
Porcine epidemic diarrhea (PED) is one of the most important diseases in many swine-raising countries. PED caused not only the death of newborn piglets, but also the weight loss in pigs of all ages due to porcine epidemic diarrhea virus (PEDV)—induced severe symptom like massive diarrhea and dehydration which resulted serious damage in the swine industry [1]. Following the PED was reported in 1978 [1], disease outbreaks have been reported in Italy [2] and United States [3], as well as China [4], Thailand [5], and Korea [6].
PEDV is an RNA virus belonging to the order Nidovirales, the family Coronaviridae, subfamily Coronavirinae, genus Alphacoronavirus [7]. It is an enveloped virus possessing an approximately 28 kb, positive sense, single-stranded RNA genome with a 5′ cap and a 3′ polyadenylated tail [8]. The genome comprised four structural proteins [spike (S), envelope (E), membrane (M), and nucleocapsid (N) protein] and four nonstructural proteins (1a, 1b, 3a, and 3b). Several viral proteins are important for inducing an immune response to PEDV. The S glycoprotein of PEDV plays an important role in induction of neutralizing antibodies, specific receptor binding, and cell membrane fusion [9]. And the S glycoprotein of PEDV possessed an immunodominant region S1 plays a crucial role in the early steps of infection [10]. The M protein plays an important role in the viral assembly process [11], also can neutralize anti-M antibody in the present of complement [12], and stimulate the production of α-interferon [13]. The N protein forms a helical nucleocapsid with genomic RNA and is the predominant antigen produced in coronavirus-infected cells, thus making it a major viral target for the accurate and early diagnosis of PEDV infection [14]. At present, several PEDV vaccines have been developed [15–17]. However, the efficacy of the available commercial vaccines is limited or the protective immunity is insufficient [18, 19]. So, it is urgently needed to develop a high-effective, rapid-acting antiviral strategy against PEDV. RNA interference (RNAi) is a natural mechanism by which double-stranded RNA directs sequence-specific degradation of messenger RNA in eukaryotic cells [20]. The use of RNAi has been successfully applied in infection inhibition studies of human and other animal viruses in vitro and in vivo, such as SARS-CoV [21], influenza virus A [22], tick-borne encephalitis virus [23], porcine transmissible gastroenteritis virus [24], and foot-and-mouth disease virus [25]. Here, we investigate whether shRNA-mediated RNAi could inhibit PEDV replication in vitro.
Materials and methods
Cell culture, virus propagation, and titration
Vero cells were maintained in Modified Eagle’s Medium (MEM) supplemented with 10 % fetal bovine serum (FBS) and cultured at 37 °C in a humidified atmosphere with 5 % CO2. The cell culture adapted PEDV strain CV777 (GenBank No. AF353511) was propagated in Vero cells as previously described [26]. After 80 % of the virus-infected cells showed cytopathic effects (CPE), the cultures were collected for three frozen-thawed cycles. Virus titration was carried out using a 96-well microplate with Vero cells. Virus cultures were 10-fold serially diluted with the virus replication medium containing trypsin (10 μg/mL). Confluent Vero cells of the microplate were washed three times with PBS and inoculated at 0.1 mL per well into five wells. Following adsorption for 1 h at 37 °C, the inocula were removed, and the cells were washed three times with PBS. Subsequently, 0.1 mL of fresh virus replication medium was transferred into each well, and the cells were further incubated for 3 days at 37 °C. Fifty percent tissue culture infective dose (TCID50) was calculated using the Reed and Muench method.
ShRNA sequences selection and siRNA expression plasmid construction
The siRNAs targeting the M, N, and S genes of PEDV were designed by Dharmacon’s siRNA design algorithm (http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx). A scrambled siRNA sequence was designed for use as a negative control. All siRNAs were synthesized by Shanghai Sangon Biotech Company (Shanghai, China). To guarantee a similar RNAi effect on different PEDV strains, the five sequences were analyzed by a BLAST search in the GenBank nucleotide database to avoid any similar sequence found in the swine genome, but share a 100 % homology within the published sequences of different PEDV strains. Their corresponding sequences are separately shown in Tables 1 and 2.
Table 1.
The inserted sequences in shRNA-expressing plasmids. The designed siRNA sequences were as follows
| Target name | siRNA sequences | Positions (bp)a |
|---|---|---|
| M1 | CTGGAATTTCACATGGAAT | 25732–25750 |
| M2 | CGTACAGGTAAGTCAATTA | 26158–26176 |
| N | CCTAAGAAGAACAAATCCA | 27154–27172 |
| S1 | CGGCATAACATGGGATAAT | 21120–21138 |
| S2 | CTGCATATGTTAATGATGA | 22745–22763 |
| NC | CTAGTATGTGCGTGCGGTT |
aIndicated the position in the whole-gene sequence of CV777 (GenBank No: AF353511)
Table 2.
The inserted sequences in shRNA-expressing plasmids. List of the siRNA target sequences
| Plasmids | Inserts |
|---|---|
| pSilencer4.1-M1 | 5′-gatccCTGGAATTTCACATGGAATTTCAAGAGAATTCCATGTGAAATTCCAGTTTTTTA-3′ |
| pSilencer4.1-M2 | 5′-gatccCGTACAGGTAAGTCAATTATTCAAGAGATAATTGACTTACCTGTACGTTTTTTA-3 |
| pSilencer4.1-N | 5′-gatccCCTAAGAAGAACAAATCCATTCAAGAGATGGATTTGTTCTTCTTAGGTTTTTTA-3′ |
| pSilencer4.1-S1 | 5′-gatccCGGCATAACATGGGATAATTTCAAGAGAATTATCCCATGTTATGCCGTTTTTTA-3′ |
| pSilencer4.1-S2 | 5′-gatccCTGCATATGTTAATGATGATTCAAGAGATCATCATTAACATATGCAGTTTTTTA-3′ |
| pSilencer4.1-NC | 5′-gatccCTAGTATGTGCGTGCGGTTTTCAAGAGAAACCGCACGCACATACTAGTTTTTTA-3′ |
The first five lowercase nucleotides represent a BamHI site (gatcc) for ligation into the pSilencer4.1-CMV vectors; the subsequent uppercase oligonucleotides represent the 19 nt sense and antisense sequences specific to the mRNA target; the italics section represents a 9 nt loop sequence common to all of the hairpins, and polythymidine tracts involved in the termination of transcription are underlined (TTTTTT)
Construction of siRNA expression plasmids is based on pSilencer4.1 (Invitrogen, Carlsbad, USA). siRNA expression plasmids were constructed by inserting annealed oligonucleotides into the BamH I and Hind III sites of pSilencer 4.1, thereby encoding the siRNA transcription unit. To investigate whether shRNA-mediated RNAi could block PEDV infection in Vero cells, all the five sequences and a non-specific sequence were generated to make shRNA-expressing plasmids: pSilencer4.1-M1, pSilencer4.1-M2, pSilencer4.1-N, pSilencer4.1-S1, pSilencer4.1-S2, or pSilencer4.1-NC, respectively.
Cell transfection and virus infection
One day before transfection, a total of 2 × 104 Vero cells in 200 μL medium without antibiotics were seeded into each well of 48-well plates. When the cells reached 70–80 % confluency, the medium was replaced and cells were transfected with 0.4 μg/well of shRNA-expressing plasmids (pSilencer4.1-M1, pSilencer4.1-M2, pSilencer4.1-N, pSilencer4.1-S1, pSilencer4.1-S2 or pSilencer4.1-NC) using the TransFast™ transfection reagent (Promega Biotech Co. Ltd., USA) according to the manufacturer’s recommendations. The cells were then incubated at 37 °C for 4 h, followed by being overlaid with MEM containing 2 % FBS (The transfection did not cause cytotoxicity for any of the shRNA transfection in the experiments). At 28 h later, the medium was replaced and the cells were challenged with PEDV at 100 TCID50, and the infection was allowed to proceed for the indicated time points. In the experiment, untransfected cells (0 μg/well) but infected with PEDV were served as the mock control. CPE were evaluated under an inverted phase contrast microscopy (Axioskop; Zeiss Co. Ltd., Oberkochen, Germany) at different time points post-infection. Cell images were captured.
MTS assay
Vero cells (1 × 104 per well) seeded in 96-well plates were treated as described in “Cell transfection and virus infection” section, the amounts of plasmids were 0.2 μg/well and the cells were challenged with PEDV at 100 TCID50 in each well. After 36 h, cell viability was assessed by adding 20 μL/well of CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS, Promega Biotech Co. Ltd., USA) to cell cultures according to the manufacturer’s instructions. The cells were incubated for 4 h with MTS, light absorbance of each solution was measured at 490 nm. Subtract the average 490 nm absorbance from the “no cell” control wells from all other absorbance values to yield corrected absorbances. The cell viability % was calculated by the treated cell OD value divide the OD value from the normal cell. The experiment was repeated three times and each performed in triplicate.
Viral titer assay
PEDV cultures in shRNA-transfected Vero cells were collected 48 h after viral infection. After three freeze—thaw cycles, the viral titer was measured using 50 % tissue culture infective dose (TCID50) assays. The cultures were serially diluted tenfold from 10−1 to 10−10, and added onto a monolayer of Vero cells in 96-well culture plates. Each dilution was added to three wells. After 3 days of infection, the TCID50 was calculated by the Reed–Muench method.
Quantitative real-time PCR
To assess the influence of shRNAs on PEDV replication, the N gene of PEDV was used as a standard for the PEDV genome. ShRNA-transfected Vero cells were harvested after 48 h of PEDV infection, and total RNA were extracted using the TaKaRa MiniBEST Universal RNA Extraction kit (Takara Bio, Dalian, China) according to the manufacturer’s instructions. These RNA temples were stored for further experiment. The levels of N mRNA transcripts were determined by qPCR using the one Step SYBR® PrimeScript™ PLUS RT-PCR Kit (Takara Bio, Dalian, China) and the following gene-specific primers: forward, 5′-CGCAAAGACTGAACCCACTAATTT-3′ and reverse, 5′-TTGCCTCTGTTGTTACTTG GAGAT-3′ for PEDV N gene [27]; forward, 5′-GGACTTCGAGCAGGAGATGG-3′, reverse, 5′-AGGAAGGAGGGCTGGAAGAG-3′ for porcine β-actin. PCR was performed in an LightCycler480 real-time detection system (Roche, USA) under the following conditions: initial reverse transcription at 42 °C for 5 min, 95 °C for 10 s; 40 cycles of PCR reaction were performed at 95 °C for 5 s, and annealing and extension at 60 °C for 30 s. Comparative threshold (Ct) values in three independent experiments were calculated by Ct method. The N mRNA level was normalized to the housekeeping gene β-actin in the same sample, and the average of 2−△△Ct value of viral RNAs [28] in each sample is represented in Fig. 4.
Fig. 4.
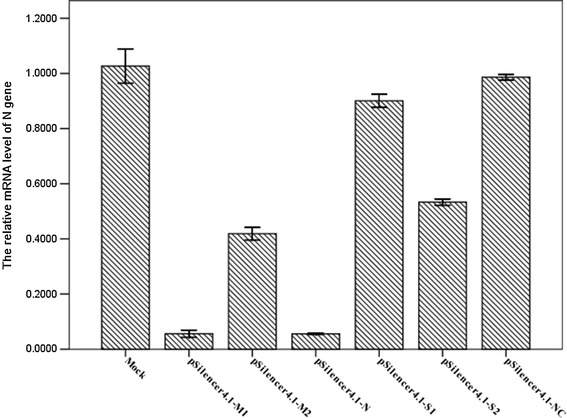
Inhibition of PEDV RNA replication by shRNAs in Vero cells. Real-time quantitative RT-PCR detection of PEDV N mRNA transcripts relative to β-actin transcripts in the same sample. The mean of three repeat experiments performed in triplicate is shown and error bars represent the SD
Statistical analysis
Data were expressed as mean ± standard deviation (SD) from three independent experiments in triplicate. Results were analyzed by Student’s t test. A p < 0.05 was considered o be statistically significant.
Results
Sequence-specific protection of Vero cells from PEDV infection by shRNAs
To investigate whether shRNAs could protect Vero cells from PEDV-induced CPE, plasmids pSilencer4.1-M1, pSilencer4.1-M2, pSilencer4.1-N, pSilencer4.1-S1, and pSilencer4.1-S2 were transfected into Vero cells, respectively. Plasmid pSilencer4.1-NC (expressing non-specific shRNA) was used as negative controls, 28 h post-transfection, Vero cells were infected with PEDV at 100 TCID50, and CPE was observed daily. At 48 h post-infection, the images of cells were captured (Fig. 1). Analysis of cell morphology indicated that Vero cells infected with virus only (Mock) or transfected with negative control plasmids (pSilencer4.1-NC) became reticulated and detached from the monolayer. However, CPE in Vero cells transfected with plasmid pSilener4.1-M1 and pSilener4.1-N expressing specific shRNAs were rarely seen. Also, plasmids pSilener4.1-S2 and pSilener4.1-M2 could potently block CPE in the cell cultures. The pSilener4.1-S1 could partially protect the Vero cells away from the PEDV-induced destruction.
Fig. 1.
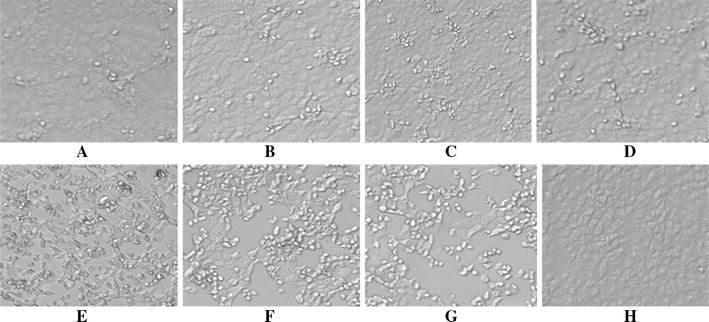
Effect of shRNAs on PEDV-induced CPE in Vero cells. Vero cells were transfected with different plasmids and then infected with PEDV at 100 TCID50. CPE were examined 48 h post-infection, and cell images were captured. From (a) to (e), cells were transfected with pSilener4.1-M1 (a), pSilener4.1-N (b), pSilener4.1-S2 (c), pSilener4.1-M2 (d), and pSilener4.1-S1 (e), respectively. f, g, and h also served as transfected with pSilener4.1-NC (f), mock control (g), and normal Vero (h) in the experiment, respectively. The experiment was performed in triplicate and repeated three times
MTS assay
To further study the effect of shRNAs on protecting Vero cells against PEDV destruction, a MTS assay was performed on the Vero cells that were transfected with shRNA-expressing plasmids, respectively, and challenged with PEDV. Because there is a linear response between cell number and absorbance at 490 nm, the absorbance at 490 nm is directly proportional to the number of living cells in culture. The mean OD values (mean ± S.D.) of solutions in wells treated with pSilener4.1-M1, pSilener4.1-M2, pSilener4.1-N, pSilener4.1-S1, and pSilener4.1-S2 were 2.059 ± 0.005, 1.902 ± 0.017, 2.065 ± 0.007, 1.233 ± 0.005, and 1.812 ± 0.014, while those treated with pSilener4.1-NC, mock, and normal were 0.32 ± 0.005, 0.326 ± 0.009, and 2.239 ± 0.011, respectively (Fig. 2a). As seen in Fig. 2b, except for the plasmid pSilener4.1-NC, all the shRNA-expressing plasmids could protect Vero cells from PEDV destruction at different levels. Plasmids pSilener4.1-M1 and pSilener4.1-N were highly effective in inhibiting the PEDV-induced CPE, compared with the mock control (p<0.05). The cell viability (%) was between 55 % and 92 %, pSilener4.1-M1 (91.961 ± 0.326 %), pSilener4.1-M2 (84.949 ± 0.586 %), pSilener4.1-N (92.259 ± 0.304 %), pSilener4.1-S1 (55.078 ± 0.151 %) and pSilener4.1-S2 (80.926 ± 0.235 %), which pSilencer4.1-NC (14.295 ± 0.138 %) and mock (14.576 ± 0.197 %).
Fig. 2.
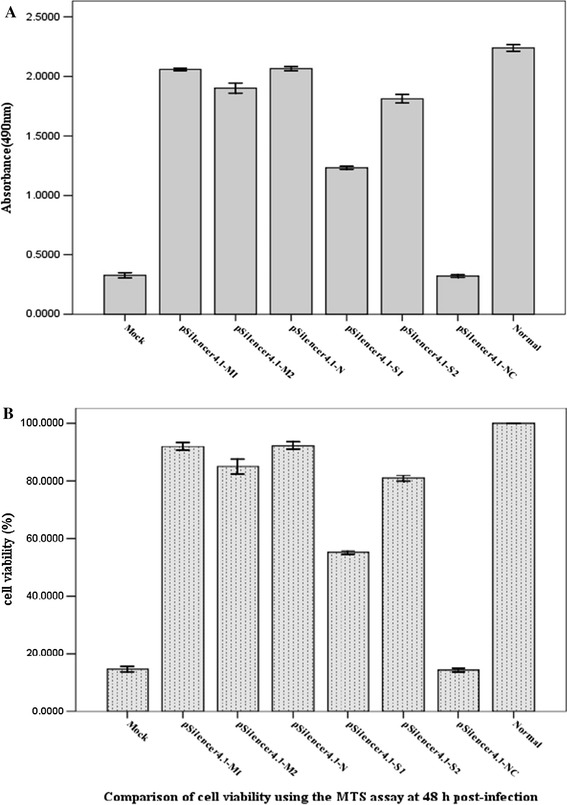
Protection of Vero cells from PEDV destruction by shRNAs. Vero cells were transfected with plasmids as indicated and then infected with PEDV at 100 TCID50. At 40 h post-infection including 4 h incubation along with MTS, viable cell numbers were evaluated by a MTS assay. OD values represent the mean ± SD of three separate experiments performed in triplicate (a). The cell viability was calculated using the MTS assay at 40 h post-infection (b)
Examination of shRNAs effect by infectious virus assay
To substantiate the inhibitory effect of shRNAs on production of viable virus, the TCID50 was used to titrate PEDV at 48 h post-infection. The results showed that in control cells transfected with pSilencer4.1-NC, the mean virus TCID50 reached 106.93 at 48 h post-infection, which was similar to that observed in mock cells (mock cells are untransfected but infected by PEDV, the mean virus TCID50 of mock group was 106.84). In contrast, the titers at 48 h post-infection were 103.46, 104.43, 103.49, 106.04, and 105.37 for cells transfected with pSilener4.1-M1, pSilener4.1-M2, pSilener4.1-N, pSilener4.1-S1, and pSilener4.1-S2, respectively (Fig. 3). The viral titers of cell lysates transfected with pSilencer4.1-M1 and pSilencer4.1-N were reduced by 1000-fold compared to that of the mock group (p < 0.05). However, the viral titers of cells transfected with pSilener4.1-S2 and pSilener4.1-M2 were reduced 10- and 100-fold (p < 0.05), respectively. These data indicated that pSilencer4.1-M1 and pSilencer4.1-N showed the maximum inhibition, whereas pSilencer4.1-M2 and pSilencer4.1-S2 showed partial virus replication inhibition while there is a small reduction in viral titers was observed but it was not statistically significant in the cells transfected with pSilener4.1-S1.
Fig. 3.
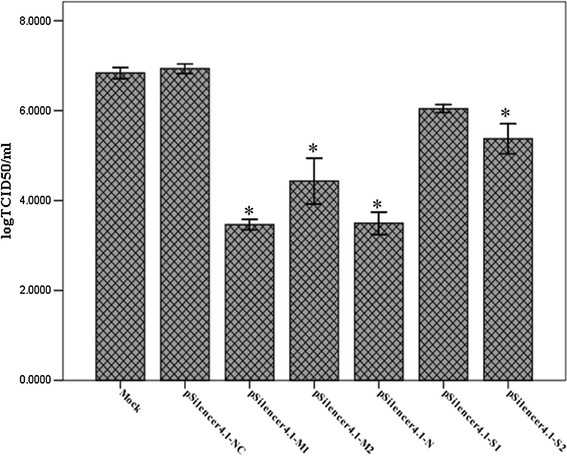
Reduction in titers of PEDV in Vero cells transfected with shRNA plasmids. Viral titers were determined by the TCID50 assay. Data are expressed as mean log TCID50/mL for each group from three separate experiments and error bars represent the SD
Effective inhibition of PEDV RNA replication by shRNAs
To assess the influence of shRNAs on PEDV replication, the N gene of PEDV was used as a standard for the PEDV genome. Then, a real-time quantitative RT-PCR analysis of N mRNA level was normalized to the corresponding β-actin in the same sample. The relative amount of N gene in mock cells was regarded as 1.000, where the relative amounts of N gene in cells infected with PEDV after being transfected with pSilencer4.1-M1, pSilencer4.1-M2, pSilencer4.1-N, pSilencer4.1-S1, pSilencer4.1-S2, and pSilencer4.1-NC were 0.056 ± 0.005, 0.418 ± 0.009, 0.055 ± 0.001, 0.780 ± 0.004, 0.533 ± 0.005, and 0.986 ± 0.004, respectively. Analysis of these data revealed that the amounts of PEDV RNAs in samples transfected with pSilencer4.1-M1 and pSilencer4.1-N were reduced by 94.4 and 94.5 %, respectively, compared to that of the mock group (p < 0.05). It suggested a potent inhibition of PEDV RNA replication triggered by sequence-specific shRNAs in Vero cells (Fig. 4).
Discussion
PEDV infection causes acute diarrhea, dehydration, and even a death rate greater than 95 % in infected piglets [29]. However, currently available vaccines and antiviral drugs are of limited value. Since 2010, PED had re-emerged in vaccinated swine herds, which leads to significant economic losses in swine husbandry in China [30–32]. RNAi is a natural cellular process mediated by small dsRNA that triggers sequence-specific gene silencing at the post-transcriptional level. RNAi can be introduced into the cells using two different approaches, exogenously introduced synthetic siRNAs or plasmid/viral vector constructs containing shRNAs. Although cells transfected with synthesized siRNAs can achieve effective silencing of a target gene, the effects are transient. The plasmid or virus vectors-based shRNAs can be cleaved by Dicer to produce siRNAs in the host cells, which can circumvent the disadvantages of chemically synthesized siRNAs using stably transfected. In terms of the fight against viral diseases, RNAi has been applied successfully to inhibit the replication of human and animal viruses using the plasmid vector constructs containing shRNAs [33–38]. In this study, we designed five siRNAs targeting various regions of N, M, and S genes of PEDV and corresponding shRNA expression plasmids were constructed. These plasmids were transfected into Vero cells; then the cells were infected with PEDV; finally, after incubation for 48 h, the antiviral activity of those plasmids in Vero cells was assessed by CPE, virus titers, MTS, and quantitative real-time PCR. The results demonstrated that the use of RNAi against PEDV has effectively inhibited the expression of the PEDV replication in vitro.
Many studies have shown that RNAi mediated by plasmids expressing shRNA could suppress virus replication when delivered into mammalian cells in vitro. It has been applied successfully to inhibit the replication of a number of viruses, such as porcine reproductive respiratory syndrome virus [39] and transmissible gastroenteritis coronavirus [37] in vitro. In this study, Vero cells transfected with the above five vectors and the negative control vectors were challenged with PEDV. Through the assessments of CPE observation, MTS assay, virus titers, and quantitative real-time PCR, the results showed that the two plasmids pSilener4.1-M1 and pSilener4.1-N, which expressing the specific shRNA targeting the PEDV gene sequences, were effectively able to inhibit the replication of PEDV. They were able to prevent Vero cells from forming CPE, and could make 1000-fold decreases of virus titers detected by TCID50. Furthermore, the results of quantitative real-time PCR showed that the two plasmids could effectively inhibit the replication of PEDV by 94.4 and 94.5 %. Now, developing the economic and effective approaches to handle PEDV is an important goal for the swine industry. Currently, vaccination, improved husbandry, and breeding pigs for improved resistance are the traditional strategies. Recently, transgenic RNAi has been developed to assist in the fight against disease. Recent research involving the phenomenon of RNAi indicates that technologies which take advantage of this evolutionary conserved process can be developed to generate transgenic animals resistant to a wide variety of different infectious diseases [40, 41]. In our study, two most effective siRNAs targeting the M and N genes in the PEDV genome were selected and corresponding shRNA expression plasmids were constructed. Also, the results demonstrated that the two plasmids could clearly inhibit the replication of PEDV in Vero cells. These findings demonstrate that RNAi could reduced viral replication in vitro significantly, thus it may be good candidate for further study of PEDV infection.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 31302101), the public research and capacity building project of Guangdong Province, China (No. 2014A010107022), the Science and Technology Planning Project of Guangdong Province, China (No. 2012B050500013), the International S&T Cooperation Program of China (No. 2014DFA31730), the Special Program of Key Research Program of Guangdong Province, China (No. 2012A020100001), the Guangzhou Science and Technology Program (No. 2013J4500031), the Special Fund for Agro-scientific Research in the Public Interest (201303046).
References
- 1.Pensaert MB, de Bouck P. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martelli P, Lavazza A, Nigrelli AD, Merialdi G, Alborali LG, Pensaert MB. Vet. Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- 3.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Liu X, Lang H, Wang Z, Shi D, Shi H, Zhang X, Feng L. Arch. Virol. 2013;158:1397–1401. doi: 10.1007/s00705-013-1608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puranaveja S, Poolperm P, Lertwatcharasakul P, Kesdaengsakonwut S, Boonsoongnern A, Urairong K, Kitikoon P, Choojai P, Kedkovid R, Teankum K, Thanawongnuwech R. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SJ, Song DS, Park BK. Arch. Virol. 2013;158:1533–1541. doi: 10.1007/s00705-013-1651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Committee on Taxonomy of Viruses, Virus taxonomy: 2012 release (2012), http://ictvonline.org/virusTaxonomy.asp?version_2012
- 8.Song D, Park B. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte M, Tobler K, Bridgen A, Rasschaert D, Ackermann M, Laude H. Virology. 1994;198:466–476. doi: 10.1006/viro.1994.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh J, Lee KW, Choi HW, Lee C. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen VP, Hogue BG. J. Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saif LJ. Vet. Microbiol. 1993;37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laude H, Gelfi J, Lavenant L, Charley B. J. Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy FA, Gibbs EP, Horzinek MC, Studdert MJ. Veterinary virology. 3. San Diego: Academic Press; 1999. pp. 496–501. [Google Scholar]
- 15.Kweon CH, Kwon BJ, Lee JG, Kwon GO, Kang YB. Vaccine. 1999;17:2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoi K, Sugioka H, Satoh T, Kadoi BK. New Microbiol. 2002;25:285–290. [PubMed] [Google Scholar]
- 17.Song DS, Yang JS, Oh JS, Han JH, Park BK. Vaccine. 2003;21:1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi HJ, Kim JH, Lee CH, Ahn YJ, Song JH, Baek SH, Kwon DH. Antivir. Res. 2009;81:77–81. doi: 10.1016/j.antiviral.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song HJ, Chae SW, Yoon KA, Park JS, Choi HJ, Cosmetic J. Public Health. 2010;6:42–44. [Google Scholar]
- 20.Hannon GJ. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 21.Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL. Antivir. Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui HY, Zhao GY, Huang JD, Jin DY, Yuen KY, Zheng BJ. PLoS One. 2009;4:5671. doi: 10.1371/journal.pone.0005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achazi K, Patel P, Paliwal R, Radonić A, Niedrig M, Donoso-Mantke O. Antivir. Res. 2012;93:94–100. doi: 10.1016/j.antiviral.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Huang F, Hua X, Cui L, Zhang W, Shen Y, Yan Y, Chen P, Ding D, Mou J, Chen Q, Lan D, Yang Z. Virus Res. 2010;149:51–55. doi: 10.1016/j.virusres.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu YF, Shen HY, Zhao MQ, Chen LJ, Li YG, Liao M, Jia JT, Lv YR, Yi L, Chen JD. J. Virol. Methods. 2012;181:51–58. doi: 10.1016/j.jviromet.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann M, Wyler R. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SH, Kim IJ, Pyo HM, Tark DS, Song JY, Hyun BH. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Kang TJ, Kim YS, Jang YS, Yang MS. Vaccine. 2005;23:2294–2297. doi: 10.1016/j.vaccine.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Fan JH, Zuo YZ, Li JH, Pei LH. Virus Genes. 2012;45:113–117. doi: 10.1007/s11262-012-0755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Yang J, Yu F, Ge J, Lin T, Song T. Virus Genes. 2014;45:499–507. doi: 10.1007/s11262-012-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li ZL, Zhu L, Ma JY, Zhou QF, Song YH, Sun BL, Chen RA, Xie QM, Bee YZ. Virus Genes. 2012;45:181–185. doi: 10.1007/s11262-012-0735-8. [DOI] [PubMed] [Google Scholar]
- 33.Peng S, York JP, Zhang P. Proc. Natl. Acad. Sci. 2006;103:2252–2256. doi: 10.1073/pnas.0511034103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 35.Yang YJ, Zhao PS, Zhang T, Wang HL, Liang HR, Zhao LL, Wu HX, Wang TC, Yang ST, Xia XZ. Virus Res. 2012;169:169–174. doi: 10.1016/j.virusres.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang F, Zhou J, Yang Z, Cui L, Zhang W, Yuan C, Yang S, Zhu J, Hua X. Vet. Microbiol. 2010;142:261–267. doi: 10.1016/j.vetmic.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L, Zhang YM, Dong LJ, Cheng M, Wang J, Tang QH, Wang G. Virol. J. 2012;9:176. doi: 10.1186/1743-422X-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Dai Y, Liu S, Guo H, Wang T, Ouyang H, Tu C. Antivir. Res. 2011;91:209–216. doi: 10.1016/j.antiviral.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao Y, Guo Y, Zhang L, Zhao Z, Li N. Mol. Biol. Rep. 2012;39:2515–2522. doi: 10.1007/s11033-011-1003-z. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Li Q, Bao Y, Li J, Chen Z, Yu X, Zhao Y, Tian K, Li N. J. Biotechnol. 2014;171:17–24. doi: 10.1016/j.jbiotec.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsoondar J, Vaught T, Ball S, Mendicino M, Monahan J, Jobst P, Vance A, Duncan J, Wells K, Ayares D. Xenotransplantation. 2009;16:164–180. doi: 10.1111/j.1399-3089.2009.00525.x. [DOI] [PubMed] [Google Scholar]


