Abstract
There are few studies involving the detection of Aichivirus B in cattle herds worldwide, and this virus has never been diagnosed in South America. This study evaluated 222 diarrhoeic faecal samples from four Brazilian geographical regions (South, Southeast, Midwest, and North), collected between February 2010 to May 2012. To evaluate the frequency of occurrence in different types of livestock, samples from beef (n = 105) and dairy (n = 117) cattle herds were evaluated. To determine the category of animals more susceptible to infection, the sampling included samples from calves (n = 182) and adults animals (n = 40). The 216 bp fragment of the Aichivirus RdRp gene was amplified by a RT-PCR assay in 18.2 % (40/222) of the samples evaluated in both beef and dairy cattle animals. The highest (P < 0.05) detection rate (20.9 %; 38/182) of the Aichivirus B was found in calves. The nucleotide sequencing analysis showed that the Brazilian Aichivirus B strains clustered in a distinct branch in the phylogenetic tree of the European and Asiatic strains. This is the first description of Aichivirus B infection in Brazilian cattle herds.
Keywords: Bovine, Diarrhoea, Dairy cattle, Beef cattle, RT-PCR, Enteric picornaviruses
Introduction
The genus Kobuvirus belongs to the Picornaviridae family, and the virions are non-enveloped, with icosahedral symmetry and a diameter of 27–30 nm. The virus genome consists of single-stranded positive-sense RNA, and its size ranges from 8.2 to 8.4 Kb (Reuter et al. 2011).
Currently, the Kobuvirus genus includes three species, designated as Aichivirus A, Aichivirus B, and Aichivirus C, which can cause infections in humans, cattle, and pigs, respectively (ICTV 2013).
Aichivirus A (A846/88 strain) was first isolated in 1989 from a patient with nonbacterial acute gastroenteritis in an outbreak in Japan. The cases were associated with consumption of oysters (Yamashita et al. 1991). Additionally, in Japan during the year 2003, Aichivirus B (U-I strain) was detected in bovine serum and faecal samples from clinically healthy cattle (Yamashita et al. 2003). In 2007, the Aichivirus C (S-1-HUN strain) was identified in a healthy piglet in Hungary (Reuter et al. 2008).
In addition to humans, cattle, and pigs, infections of the kobuvirus have been described in other animal species such as dogs (Li et al. 2011; Kapoor et al. 2011), bats (Li et al. 2010), wild boar (Reuter et al. 2013), and sheep (Reuter et al. 2010). Reuter et al. (2011) believe that there is a high possibility of kobuvirus also infecting wild animals.
There are few studies involving the detection of Aichivirus B in cattle. Analyzing a small number of the faecal samples (n = 9), Barry et al. (2011) reported a high infection rate of Aichivirus B in the Netherlands. In Thailand, this virus has been detected in diarrhoeic faecal samples of calves between 7 and 49 days of age (Khamrin et al. 2008). In Hungary, the Aichivirus B was also described by Reuter and Egyed (2009) in adult cattle. In South Korea, Park et al. (2011) analysed faecal samples from cattle with diarrhoea, and detected higher presence of Aichivirus B in calves than in adults.
Jeoung et al. (2011) identified this virus in faecal samples of symptomatic and asymptomatic calves (≤30-day-old). In Europe, epidemiological studies were conducted in animals with (Mauroy et al. 2009) and without (Di Martino et al. 2012) diarrhoea, showing the presence of Aichivirus B.
The kobuvirus is considered a new enteric virus, and the clinical consequences of the infection caused by this agent are not yet established in Brazil. In this study, we investigated the prevalence of the Aichivirus B in faecal samples of Brazilian cattle herds and the relationship of Aichivirus B with other viruses that can cause diarrhoea in cattle.
Materials and methods
Inclusion criteria
A total of 222 diarrhoeic faecal samples were collected between February 2010 and May 2012. The sampling was representative of the four main geographical regions of Brazil with high numbers of bovine livestock (South, Southeast, Midwest, and North). As a secondary inclusion criterion, faecal samples from beef (n = 105) and dairy (n = 117) cattle herds were selected to evaluate the frequency of kobuvirus infection in different types of livestock. Finally, to determine the category of animals most susceptible to infection, diarrhoeic faecal samples from young (calves ≤ 60 days old, n = 182) and adult animals (≥ 1 year old, n = 40) were selected. Table 1 shows the distribution of diarrhoeic faecal samples included in this study according to the origin, rearing system of the farms, and age of the animals.
Table 1.
Distribution of diarrhoeic faecal samples by geographical location, rearing system of the farms and age of the animals included in the study for Aichivirus B detection by RT-PCR
| Origin | Sampling (n) | ||||||
|---|---|---|---|---|---|---|---|
| Region | State | Herd | Samples | ||||
| Dairy | Beef | Calves | Adult | Dairy | Beef | ||
| South | Paraná | 22 | 4 | 83 | 37 | 75 | 45 |
| Southeast | Minas Gerais | 1 | - | 20 | - | 20 | - |
| São Paulo | 1 | - | 6 | - | 6 | - | |
| Midwest | Goiás | 3 | - | 13 | 3 | 16 | - |
| Mato Grosso | - | 1 | 20 | - | - | 20 | |
| Mato Grosso do Sul | - | 1 | 5 | - | - | 5 | |
| North | Rondônia | 2 | - | 15 | - | - | 15 |
| Pará | - | 1 | 20 | - | - | 20 | |
| Total | 29 | 7 | 182 | 40 | 117 | 105 | |
RNA extraction
Faecal suspensions were prepared at 10–20 % (w/v) in 0.01 M phosphate-buffered saline (PBS) pH 7.2 (137 mM NaCl; 3 mM KCl; 8 mM Na2HPO4; 14 mM KH2PO4), and centrifuged at 3,000×g for 5 min. The RNA extraction was performed with 400 μL of faecal suspensions using a combination of phenol/chloroform/isoamyl alcohol (25:24:1) and silica/guanidinium isothiocyanate methods as described by Alfieri et al. (2006). The RNA was eluted in 50 μL of ultra-pure RNase-free DEPC-treated sterile water. Sterile water was included as a negative control in all viral RNA extraction procedures.
Virological analysis
The RT-PCR assay to detect the kobuvirus was performed using the primers UNIV-kobu-F/R that were designed based on Kobuvirus strains, which target a 216 bp region of the RdRp gene (Reuter et al. 2009).
The SN-PCR assay for the bovine coronavirus was performed according to Takiuchi et al. (2006) to amplify a sequence based on the highly conserved region from the Mebus strain N gene (GenBank accession number U00735) with a predicted product of 251 bp.
The RT-PCR and SN-PCR products were analysed by electrophoresis in a 2 % agarose gel in TBE buffer pH 8.4 (89 mM Tris; 89 mM boric acid; 2 mM EDTA), stained with ethidium bromide, and visualized under UV light.
The presence of segmented double-stranded RNA (dsRNA) of rotavirus in the diarrhoeic faecal samples was evaluated by polyacrylamide gel electrophoresis (PAGE) technique (Pereira et al. 1983) followed by silver staining according to Herring et al. (1982).
Sequencing and phylogenetic analysis
One RT-PCR positive faecal sample with better quality of each positive herd (n = 21) was submitted to sequencing analysis to confirm the presence of Aichivirus B. RT-PCR products were purified using the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK), quantified with a Qubit® Fluorometer (Invitrogen Life Technologies, Eugene, OR, USA), and sequenced in an ABI3500 Genetic Analyser sequencer using the forward and reverse primers with the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Sequence quality analysis was carried out using Phred software, and the contig assembly was obtained using the CAP3 software (http://asparagin.cenargen.embrapa.br/phph/). Sequence similarities were performed using the BLAST software (http://blast.ncbi.nlm.nih.gov/), to verify the nucleotide similarity with sequences that are deposited in GenBank. Four nucleotide sequences with quality were selected to perform the phylogenetic tree and the nucleotide matrix identity using the MEGA version 5.10 and Bioedit version 7.0.5.3 software, respectively. The analyses were based on the neighbor-joining method from the Tamura-Nei model. Bootstrapping was statistically supported with 1,000 replicates. The referenced sequences included in this study were acquired from the National Center for Biotechnology Information, USA (GenBank) (http://www.ncbi.nlm.nih.gov/GenBank/).
Statistical analysis
Statistical analysis was performed with EpiInfo™ to compare the proportions of Aichivirus B positive samples among two categories: animal age (calves x adult) and cattle type (dairy x beef). The analysis was performed using the Chi-square (χ2) test and Fisher’s exact test. The confidence limit for the statistical tests was set at 95 % (P < 0.05).
Results
A product of 216 bp from the kobuvirus RdRp gene was amplified in 18.2 % (40/222) of the diarrhoeic faecal samples. Aichivirus B positive faecal samples originated from Brazilian geographical regions, and the states that were evaluated are shown in Table 2.
Table 2.
Aichivirus B positive diarrhoeic faecal samples from beef and dairy cattle in the four Brazilian geographical regions
| Geographic regiona | Samples Positive/ Total |
Total (%) | |
|---|---|---|---|
| Beef | Dairy | ||
| South | 4/45 | 19/75 | 23/120 (19.2) |
| Southeast | - | 6/26 | 6/26 (23.1) |
| Midwest | 3/25 | 1/16 | 4/41 (9.8) |
| North | 7/35 | - | 7/35 (20.0) |
aStates: South (Paraná); Southeast (Minas Gerais and São Paulo); Midwest (Goiás, Mato Grosso, and Mato Grosso do Sul); North (Pará and Rondônia)
Table 3 shows the distribution of diarrhoeic faecal samples that were positive for Aichivirus B collected from beef and dairy cattle herds and from calves and adult animals. The frequency of positive faecal samples from calves was statistically higher than in adult animals (P = 0.0367).
Table 3.
Detection of Aichivirus B by RT-PCR assay in diarrhoeic faecal samples of calves and adult animals in the Brazilian beef and dairy cattle herds
| Age group | Cattle herds | Animals evaluated | ||||
|---|---|---|---|---|---|---|
| Beef | Dairy | |||||
| Total | Positives (%) | Total | Positive (%) | Total | Positive (%) | |
| Calves | 68 | 13 (19.1c) | 114 | 25 (21.9c) | 182 | 38 (20.9a)* |
| Adult | 37 | 1 (2.7) | 3 | 1 (33.3) | 40 | 2 (5.0b)* |
| Total | 105 | 14 (13.3d) | 117 | 26 (22.2d) | 222 | 40 (18.2) |
*The same letter in the same line indicates non-significant values and different letters in different lines indicates significant values (P < 0.05)
The 21 RT-PCR amplicons were sequenced and confirmed the presence of Aichivirus B RdRp gene. Four Aichivirus B nucleotide sequences from calves (≤60 days old) with better quality were selected to perform the phylogenetic analysis. The four sequences (BRA09/2011, BRA1682/2011, BRA02/2012, and BRA55/2012) analysed in this study formed a new branch in the phylogenetic tree (Fig. 1). The BRA09/2011 strain showed high nucleotide identity (93.7 %) with the Hungary strain (FJ225406), while the BRA1682/2011, BRA02/2012, and BRA55/2012 strains presented high nucleotide identity (91.6 % to 92.1 %; 93.7 % to 94.2 %; 94.2 %) with the Thailand strains (EF659450, EF659451, and EF659454), respectively. The gene sequences described in the present study have been deposited in the GenBank database under accession numbers KC921389, KC921390, KC921391, and KJ402443.
Fig. 1.
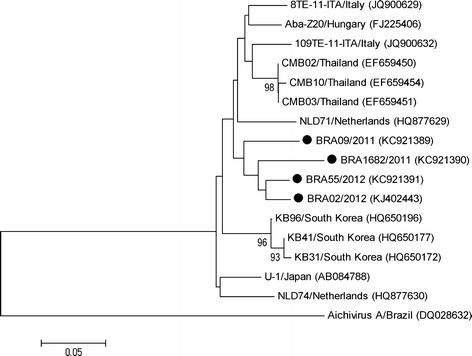
Phylogenetic analysis of a partial nucleotide sequence (nt 192) of the RdRp gene of Aichivirus B. The tree was generated using the neighbor-joining method and Tamura-Nei as the nucleotide substitution model. Bootstraps values (1,000 replicates) higher than 70 % are shown. The Brazilian strains of Aichivirus B described in this study are marked with a filled circle. Brazilian Aichivirus A strain was used as an outgroup
The presence of co-infection with other enteropathogenic virus was found only in dairy production herds. Five of the 38 Aichivirus B positive faecal samples of calves presented co-infection with other enteric viruses, including the bovine group A rotavirus (n = 2) and bovine coronavirus (n = 3).
Discussion
The RNA of Aichivirus B was detected in all Brazilian regions included in this study and represents the first detection of this virus. Similar rates of Aichivirus B infection were detected in the three regions; South (19.17 %), Southeast (23.1 %), and North (20 %). The Midwest region showed the lowest detection rate (9.76 %). These results suggest that the infection is disseminated in Brazilian beef and dairy cattle herds.
Aichivirus B was detected in 58 % (21/36) of the cattle herds evaluated (data not shown), and all regions included in the survey had at least one infected herd. The Parana State demonstrated the highest rate of virus detection, with 50 % of herds testing positive, which shows that Aichivirus B infection in cattle is widespread among herds in this state. Comparing the two types of livestock production, the highest (22.2 %; 26/117) detection rate of Aichivirus B positive diarrhoeic faecal samples was found in dairy herds, while in beef cattle herds the virus was found in 13.3 % (14/105) of the evaluated samples. However, there was no statistically significant difference (P > 0.05) between the presence of infection in dairy and beef herds.
Aichivirus B has been detected in Japan (Yamashita et al. 2003), Thailand (Khamrin et al. 2008), Belgium (Mauroy et al. 2009), Netherlands (Barry et al. 2011), Hungary (Reuter and Egyed 2009), South Korea (Jeoung et al. 2011; Park et al. 2011) and Italy (Di Martino et al. 2012) but there are no studies regarding the detection of this virus in Brazilian dairy and/or beef cattle herds. In Thailand, Khamrin et al. (2008) detected 8.3 % (6/72) of faecal samples positive for the Aichivirus B. The present study found 18 % (40/222) of faecal samples positive for this enteric picornavirus. Studies in other countries describe the presence of Aichivirus B as a common enteric pathogen of cattle. The virus was present in 4.9 % (7/142), 37.2 % (32/86), 77.8 % (7/9) of the faecal samples evaluated in Italy (Di Martino et al. 2012), South Korea (Jeoung et al. 2011), and the Netherlands (Barry et al. 2011), respectively. In this study Aichivirus B was detected in 20.9 % (38/182) of the faecal samples from calves ≤60 days of age, while studies performed in South Korea found the Aichivirus B in 40.9 % and 66.7 % of samples from calves (Jeoung et al. 2011; Park et al. 2011). The high prevalence of infection in young animals can be explained by the fact that this virus belongs to the group of agents responsible for neonatal disease, which has also been observed in pigs, and the highest positive rate for Aichivirus C is found in piglets (Park et al. 2010; Ribeiro et al. 2013; Verma et al. 2013).
There are few studies analysing the presence of Aichivirus B in adult animals with diarrhoea. In this report, Aichivirus B infection was more frequent (P < 0.05) in calves than in adult animals (Table 2). In Hungary, Reuter and Egyed (2009) described that 6.25 % (2/32) of the 1-year-old animals were positive for Aichivirus B. In South Korea, Park et al. (2011) showed that 18.4 % (9 out of 49) of the cows evaluated were positive for Aichivirus B. In the present study, this virus was identified only in 5 % (2/40) of diarrhoeic faecal samples from animals older than 1 year. In addition to cattle, in pigs the percentage of the positive animals decreased with increasing age (An et al. 2011; Ribeiro et al. 2013).
The highest rate of positive results of Aichivirus B is found in animals with clinical signs of diarrhoea (Jeoung et al. 2011; Di Martino et al. 2012). The presence of gastroenteritis that is caused by Aichivirus has been reported by Park et al. (2011) who found a high frequency (25.8 %; 16/62) of positive faecal samples from diarrhoeic animals. However, some studies have described the Aichivirus B in asymptomatic animals (Reuter and Egyed 2009; Di Martino et al. 2012).
The sequencing analysis of four amplicons confirmed the presence of Aichivirus B RNA in the faecal samples. The Brazilian Aichivirus B strains formed a distinct branch in the phylogenetic tree compared with the sequences belonging to other countries (Hungary, Japan, Thailand, Italy, South Korea, and the Netherlands).
In humans, isolates of Aichivirus have been found only in patients with gastroenteritis, which has also been described in swine, and there are studies in cattle describing the presence of this virus in cases of gastroenteritis (Yamashita et al. 2003; Jeoung et al. 2011; Ribeiro et al. 2013).
In this study, the presence of co-infection with other viral agents responsible for causing diarrhoea in cattle was observed in only five (13.2 %; 5/38) positive Aichivirus B diarrhoeic faecal samples of calves. This result agrees with the study realised by Jeoung et al. (2011), in which low Aichivirus B interaction with other agents was demonstrated in the faecal samples of the cattle that were evaluated.
However, the involvement of Aichivirus B as an enteropathogen is not yet well elucidated. Experimental infections in gnotobiotic animals are necessary to determine the involvement of this virus as an enteric disease in cattle.
In conclusion, this is the first report of detection of Aichivirus B in Brazilian beef and dairy cattle herds. The virus was detected in all four geographical regions and eight states included in this study, suggesting that it is distributed across the country. The frequency of infection in diarrhoeic animals is higher in calves than in adults.
Acknowledgements
This study was supported by the following Brazilian institutes: National Council of Scientific and Technological Development (CNPq), Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES), Financing of Studies and Projects (FINEP), and the Araucária Foundation (FAP/PR). Alfieri, A.A., Alfieri, A.F. and Lorenzetti, E. are recipients of CNPq fellowships.
References
- Alfieri AA, Parazzi ME, Takiuchi E, Medici KC, Alfieri AF. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998–2002. Trop Anim Health Prod. 2006;38(7–8):521–526. doi: 10.1007/s11250-006-4349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An DJ, Jeoung HY, Jeong W, Lee HS, Park JY, Kim B. Porcine kobuvirus from pig stool in Korea. Virus Genes. 2011;42(2):208–211. doi: 10.1007/s11262-010-0561-9. [DOI] [PubMed] [Google Scholar]
- Barry AF, Ribeiro J, Alfieri AF, van der Poel WH, Alfieri AA. First detection of kobuvirus in farm animals in Brazil and the Netherlands. Infect Genet Evol. 2011;11(7):1811–1814. doi: 10.1016/j.meegid.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Di Martino B, Di Profio F, Di Felice E, Ceci C, Pistilli MG, Marsilio F. Molecular detection of bovine kobuviruses in Italy. Arch Virol. 2012;157(12):2393–2396. doi: 10.1007/s00705-012-1439-z. [DOI] [PubMed] [Google Scholar]
- Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16(3):473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV - International Committee on Taxonomy of Viruses (2013) http://www.ictvonline.org/virusTaxonomy.asp?bhcp=1. Accessed 25 Jan 2013
- Jeoung HY, Lim JA, Jeong W, Oem JK, An DJ. Three clusters of bovine kobuvirus isolated in Korea, 2008–2010. Virus Genes. 2011;42(3):402–406. doi: 10.1007/s11262-011-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Dubovi EJ, Qaisar N, Henriquez JA, Medina J, Shields S, Lipkin WI. Characterization of a canine homolog of human Aichivirus. J Virol. 2011;85(21):11520–11525. doi: 10.1128/JVI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamrin P, Maneekarn N, Peerakome S, Okitsu S, Mizuguchi M, Ushijima H. Bovine kobuviruses from cattle with diarrhea. Emerg Infect Dis. 2008;14(6):985–986. doi: 10.3201/eid1406.070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84(14):6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92(Pt 11):2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauroy A, Scipioni A, Mathijs E, Thys C, Thiry E. Molecular detection of kobuviruses and recombinant noroviruses in cattle in continental Europe. Arch Virol. 2009;154(11):1841–1845. doi: 10.1007/s00705-009-0518-2. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kim HK, Moon HJ, Song DS, Rho SM, Han JY, Nguyen VG, Park BK. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch Virol. 2010;155(11):1803–1811. doi: 10.1007/s00705-010-0774-1. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kim HK, Song DS, Moon HJ, Park BK. Molecular detection and genetic characterization of kobuviruses in fecal samples collected from diarrheic cattle in Korea. Infect Genet Evol. 2011;11(5):1178–1182. doi: 10.1016/j.meegid.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Pereira HG, Azeredo RS, Leite JP, Candeias JA, Racz ML, Linhares AC, Gabbay YB, Trabulsi JR. Electrophoretic study of the genome of human rotaviruses from Rio de Janeiro, Sao Paulo and Para, Brazil. J Hyg (Lond) 1983;90(1):117–125. doi: 10.1017/S0022172400063919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Egyed L. Bovine kobuvirus in Europe. Emerg Infect Dis. 2009;15(5):822–823. doi: 10.3201/eid1505.081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Boldizsar A, Kiss I, Pankovics P. Candidate new species of Kobuvirus in porcine hosts. Emerg Infect Dis. 2008;14(12):1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Boldizsar A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154(1):101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- Reuter G, Boros A, Pankovics P, Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis. 2010;16(5):869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Boros A, Pankovics P. Kobuviruses - a comprehensive review. Rev Med Virol. 2011;21(1):32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- Reuter G, Nemes C, Boros A, Kapusinszky B, Delwart E, Pankovics P. Porcine kobuvirus in wild boars (Sus scrofa) Arch Virol. 2013;158(1):281–282. doi: 10.1007/s00705-012-1456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro J, de Arruda LR, Alfieri AF, Alfieri AA. High frequency of Aichivirus C (porcine kobuvirus) infection in piglets from different geographic regions of Brazil. Trop Anim Health Prod. 2013 doi: 10.1007/s11250-013-0428-x. [DOI] [PubMed] [Google Scholar]
- Takiuchi E, Stipp DT, Alfieri AF, Alfieri AA. Improved detection of bovine coronavirus N gene in faeces of calves infected naturally by a semi-nested PCR assay and an internal control. J Virol Methods. 2006;131(2):148–154. doi: 10.1016/j.jviromet.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma H, Mor SK, Abdel-Glil MY, Goyal SM. Identification and molecular characterization of porcine kobuvirus in U. S. swine. Virus Genes. 2013;46(3):551–553. doi: 10.1007/s11262-013-0879-1. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164(5):954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84(Pt 11):3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]


