Abstract
Porcine deltacoronavirus (PDCoV) is an emerging swine enteropathogenic coronavirus that causes watery diarrhea, vomiting and mortality in newborn piglets. Previous studies have suggested that PDCoV infection antagonizes RIG-I-like receptor (RLR)-mediated IFN-β production to evade host innate immune defense, and PDCoV-encoded nonstructural protein nsp5 and accessory protein NS6 are associated with this process. However, whether the structural protein(s) of PDCoV also antagonize IFN-β production remains unclear. In this study, we found that PDCoV nucleocapsid (N) protein, the most abundant viral structural protein, suppressed Sendai virus (SEV)-induced IFN-β production and transcription factor IRF3 activation, but did not block IFN-β production induced by overexpressing RIG-I/MDA5. Furthermore, study revealed that PDCoV N protein interacted with RIG-I and MDA5 in an in vitro overexpression system and evident interactions between N protein and RIG-I could be detected in the context of PDCoV infection, which interfered with the binding of dsRNA and protein activator of protein kinase R (PACT) to RIG-I. Together, our results demonstrate that PDCoV N protein is an IFN antagonist and utilizes diverse strategies to attenuate RIG-I recognition and activation.
Keywords: Porcine deltacoronavirus, Nucleocapsid, Interferon, RIG-I, RNA binding
Introduction
The family Coronaviridae contains four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus [1, 2]. Deltacoronavirus is a newly identified genus and to date, only one deltacoronavirus, porcine deltacoronavirus (PDCoV), has been successfully isolated by cell culture in vitro [3]. PDCoV was first detected in pig samples in 2012 in Hong Kong [2]. However, the clinical significance of PDCoV was not addressed until the emergence of PDCoV in 2014 in Ohio, USA, when it rapidly spread within the country to at least 20 states [4–6]. Thereafter, Canada, South Korea, Mainland China, Thailand, Lao People’s Democratic Republic and Vietnam reported the emergence of PDCoV [7–11]. Animal experiments have shown that PDCoV infection mainly causes severe atrophic enteritis, accompanied by severe diarrhea, vomiting and death [12]. PDCoV may also transmit between species and a recent study proved that calves are susceptible to PDCoV, posing a significant threat to animal health [13].
The full-length genome of PDCoV is approximately 25.4 kb in length with the essential genes occurring in the order 5′ UTR-ORF1a/1b-S-E-M-NS6-N-NS7-NS7a-3′ UTR. The genome is believed to encode 15 mature nonstructural proteins (nsps), four structural proteins and three accessory proteins [8]. The nucleocapsid (N) protein is the most abundant viral protein in both infected cells and virions and exhibits multiple functions throughout the life cycle of the virus [14]. In addition to maintaining the viral RNA in an ordered conformation for replication and transcription [15–18], and being involved in the regulation of inflammatory cytokines [19, 20], RNA interference [21] and apoptosis [22], N protein is also responsible for counteracting the host innate immune defense. Previous studies have revealed that N proteins from porcine epidemic diarrhea virus (PEDV), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and mouse hepatitis virus A59 (MHV-A59) are involved in suppressing IFN-β production. PEDV N protein blocks IFN-β production by targeting TANK-binding kinase 1 (TBK1) to prevent its interaction with IRF3 [23], while MHV and SARS-CoV N proteins exert IFN-antagonizing function by targeting protein activator of the interferon-induced protein kinase (PACT) and impairing PACT-RIG-I/MDA5 interactions [24]. SARS-CoV and MERS-CoV N proteins have also been reported to antagonize IFN-β production by interacting with tripartite motif protein 25 (TRIM25) and other unidentified mechanisms [25]. Until now, only N proteins from alphacoronavirus and betacoronavirus have been well studied and whether the N protein of the deltacoronavirus PDCoV antagonizes type I IFN production remains unclear.
Here, we identified PDCoV N protein as an IFN-β antagonist and analyzed the possible mechanisms of action. We found that PDCoV N protein interacts with PACT, poly(I:C) and the helicase domain and C-terminal domain (CTD) of RIG-I, thus impairing the association of RIG-I with dsRNA or/and PACT. This is a distinct mechanism from N proteins of alphacoronavirus and betacoronavirus.
Materials and methods
Cells, viruses, and reagents
HEK-293T cells were cultured and maintained in RPMI-1640 (HyClone, UT), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (PAN-biotech, Bavaria, Germany) at 37 °C in a humidified 5% CO2 incubator. The PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) used in this study was isolated from a suckling piglet with acute diarrhea in China in 2014 [26]. LLC-PK1 cells, a porcine kidney cell line purchased from the ATCC, were cultured at 37 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% heat-inactivated FBS and were used to amplify PDCoV. SEV was acquired from the Centre of Virus Resource and Information at the Wuhan Institute of Virology. Recombinant VSV-GFP was generously provided by Prof. Zhigao Bu at Harbin Veterinary Research Institute, China. Poly(I) and poly(C)-agaroses were bought from Sigma-Aldrich (MO, USA). Mouse monoclonal antibodies against Flag, HA, Myc and β-actin were purchased from Medical & Biological Laboratories (Nagoya, Japan). The PDCoV-N-protein-specific monoclonal antibody was produced in our laboratory as described previously [27]. Rabbit polyclonal antibodies directed against Flag were bought from ABclone (Wuhan, China). Alexa Fluor 488-conjugated donkey anti-mouse and 594-conjugated donkey anti-rabbit antibodies were purchased from Santa Cruz Biotechnology (CA, United States).
Plasmids
The luciferase reporter plasmids IFN-β–Luc, 4 × PRDIII/I–Luc (referred to as IRF-3–Luc), and 4 × PRDII–Luc (referred to as NF-κB–Luc), and molecules in the RLR-signaling pathway including RIG-I/IN, MDA5 have been described previously [23]. pCAGGS-HA/Flag/Myc-PDCoV-N were constructed by standard RT-PCR assays with specific primers to amplify the cDNA of PDCoV N gene from the total RNA extracted from PDCoV-infected LLC-PK1 cells. Truncated mutants pCAGGS-Flag-2*CARD/Hel/CTD were constructed from RIG-I expression plasmids. All primers used in these constructs are listed in Table 1 and all constructs were validated by DNA sequencing.
Table 1.
Primers for gene amplification
| Names | Primers (5′–3′) |
|---|---|
| PDCoV-N-F | ACTGAATTCATGGCTGCACCAGTAGTCCCTAC |
| PDCoV-N-R | CTAATCGATCTACGCTGCTGATTCCTGCTTTAT |
| Flag-h-RIG-I-F | TAAATCGATATGACCACCGAGCAGCGA |
| Flag-h-RIG-I-R | CAGCTCGAGTCATTTGGACATTTCTGC |
| Flag-h-RIG-IN-R | CAGCTCGAGTCATGGACATGAATTCTC |
| Flag-h-RIG-I-CTD-F | TAAATCGATGAAAATAAAAAACTGCTCTG |
| Flag-h-RIG-I-HEL-F | TAAATCGATCCTTCAGAAGTGTCTGATA |
| Flag-h-RIG-I-HEL-R | CAGCTCGAGCTTATCAGGGACAGGTTTTGG |
Luciferase reporter gene assay
HEK-293T or LLC-PK1 cells grown in 24-well plates were co-transfected with 0.1 μg/well reporter plasmid (IFN-β–Luc, IRF-3–Luc, NF-κB–Luc), 0.02 μg/well pRL–TK plasmid (Promega), and the indicated expression plasmid or an empty control plasmid. Where indicated, the cells were also mock-infected/treated or infected/treated with SEV (ten hemagglutinating activity units/well)/poly(I:C) at 24 h after co-transfection. The cells were lysed 12 h later and the firefly luciferase and Renilla luciferase activities were determined with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Data are expressed as the relative firefly luciferase activities normalized to the Renilla luciferase activities and are representative of three independently conducted experiments.
RNA extraction and quantitative real-time RT-PCR
To determine the effects of PDCoV N protein on the expression of IFN-β, HEK-293T cells in 24-well plates were transfected with 1 μg of empty vector or plasmid encoding PDCoV N protein. After 24 h, the cells were mock-infected or infected with SEV for 12 h. Total RNA was extracted from the cells with RNA-Solv Reagent (Omega, GA) and an aliquot (1 μg) was reverse transcribed to cDNA using AMV reverse transcriptase (Roche, Basel, Switzerland). The cDNA (1 μl of the 20 μl RT reaction) was then used as the template in a SYBR Green PCR assay (Applied Biosystems). The abundance of individual mRNA in each sample was assayed three times and normalized to that of the internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Primers (IFN-β: tctttccatgagctacaacttgct/gcagtattcaagcctcccattc; GAPDH: tcatgaccacagtccatgcc/ggatgaccttgcccacagcc) were designed using Primer Express software v.3.0 (Applied Biosystems).
ELISA for IFN-β
To measure secreted IFN-β, HEK-293T cells were mock transfected or transfected with 1 μg of the expression plasmid encoding PDCoV N protein. At 24 h post-transfection, cells were mock infected or infected with SEV for 16 h. The supernatants were then harvested for ELISA with a commercial sandwich kit (PBL Assay Science) according to the manufacturer’s instructions.
Co-immunoprecipitation and immunoblotting analyses
For the transient transfection experiments, HEK-293T cells were transfected with appropriate plasmids for 28 h. The transfected cells were lysed in 200 μL of lysis buffer [4% SDS, 3% DTT, 0.065 mM Tris–HCl, (pH 6.8), 30% glycerine] supplemented with protease inhibitor (PMSF, Sigma-Aldrich). The lysates were boiled at 100 °C for 10 min before separation by SDS-PAGE and electroblotting onto a polyvinylidene difluoride membrane (Bio-Rad). Membranes were analyzed by immunoblotting with the indicated antibodies. For co-immunoprecipitation analysis, cells were washed with PBS and lysed for 20 min at 4 °C in lysis buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 10% glycerin, 0.1% SDS, and 2 mM Na2EDTA. The lysates were then cleared by centrifugation, and the proteins were immunoprecipitated overnight at 4 °C with affinity antibodies and protein A + G agarose beads (Beyotime, Shanghai, China). The immunoprecipitates were washed three times with 1 mL of lysis buffer and then analyzed by standard immunoblotting procedures.
RNA-binding assays
We assessed the RNA-binding ability by poly(I:C)-agarose-binding assay as described previously [28]. Poly(I:C)-coated agarose was made by incubating poly(C)-coated agarose with poly(I) at 56 °C for 30 min, then cooling down to 4 °C. Cell lysates were incubated with poly(I:C)-coated agarose for 1 h at 4 °C, then washed three times with lysis buffer and subjected to immunoblot analysis.
Statistical analysis
Data are presented as means and standard deviations (SD). Values of P < 0.05 were considered statistically significant, and P values of < 0.001 were considered highly statistically significant.
Results
PDCoV N protein blocks Sendai virus-induced IFN-β production
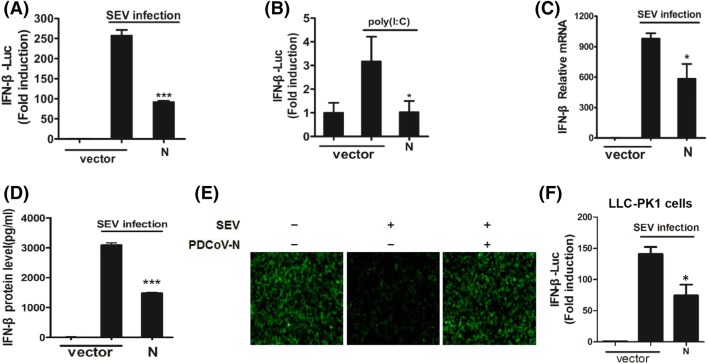
To determine whether PDCoV N protein has an inhibitory effect on IFN-β production, HEK-293T cells were co-transfected with a DNA expression construct pCAGGS-HA-PDCoV-N encoding PDCoV N protein and luciferase reporter plasmid IFN-β–Luc, together with the internal control plasmid pRL–TK, and then infected with SEV or treated with poly(I:C). As shown in Fig. 1a and b, overexpression of PDCoV N protein blocked SEV/poly(I:C)-induced IFN-β promoter activities. To confirm the IFN-β inhibitory role played by PDCoV N protein, relative levels of IFN-β mRNA in cells transfected with pCAGGS-HA-PDCoV-N and infected with SEV was measured by quantitative real-time RT-PCR (Fig. 1c). Meanwhile, supernatants were harvested for ELISA to detect secreted IFN-β (Fig. 1d). Data showed that the transcription and secretion of IFN-β were significantly reduced in PDCoV N protein-expressing cells (Fig. 1c and d). Since vesicular stomatitis virus (VSV) is sensitive to SEV-induced IFN responses, we observed the proliferation of a recombinant VSV encoding green fluorescent protein (VSV-GFP) after HEK-293T cells were transfected or mock-transfected with pCAGGS-HA-PDCoV-N prior to subsequent infection with SEV and VSV-GFP. A dramatic reduction in VSV-GFP proliferation was observed after SEV infection, but a robust restorative effect was observed in N protein-expressing cells (Fig. 1e). These results further confirmed that PDCoV N protein can antagonize IFN-β production. We repeated the luciferase assay in LLC-PK1 cells, a porcine kidney cell line supporting PDCoV infection. Consistent with the data in HEK-293T cells, PDCoV N blocked activity of the SEV-induced IFN-β promoter in LLC-PK1 cells (Fig. 1f).
Fig. 1.
PDCoV N protein suppresses SEV- or poly(I:C)-induced IFN-β production. a, b HEK-293T cells were co-transfected with IFN-β–Luc, pRL–TK plasmid and increasing concentrations of PDCoV N expression plasmids for 24 h, and then infected with SEV or treated with poly(I:C). Dual-luciferase reporter assays were performed to determine activation of the IFN-β promoter. The results represent the means and standard deviations of three independent experiments. The relative firefly luciferase activity was normalized to the Renilla reniformis luciferase activity, and the untreated empty vector control value was set as 1. c–e HEK-293T cells were transfected with mock vectors or PDCoV-N expression vectors. At 24 h post-transfection, cells were infected with SEV for 16 h. c Cells were lysed to extract total RNA, which was used for detecting the expression of IFN-β mRNA and GAPDH genes by quantitative real-time RT-PCR. The results are expressed as increases in IFN-β mRNA levels relative to those in cells transfected without SEV infection and were normalized to the expression of the GAPDH housekeeping gene. d The harvested supernatants were used to detect IFN-β by ELISA. e Cells were re-infected with VSV-GFP for 24 h, followed by analysis for fluorescence by microscopy. f Dual-luciferase reporter assays were performed in LLC-PK1 cells as described in (A). *P < 0.05; ***P < 0.001
PDCoV N protein impairs the activation of IRF3
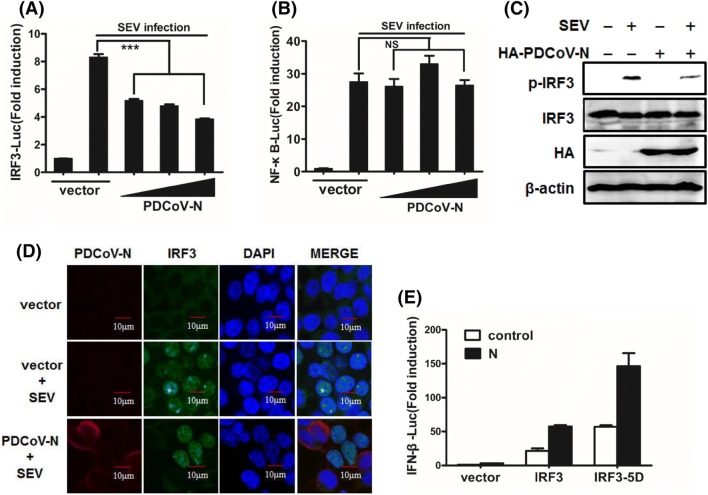
Transcription factors IRF3 and NF-κB are coordinated and cooperative mediating type I IFN production [29, 30]. To investigate whether PDCoV N protein impairs the activation of IRF3 and NF-κB, a luciferase assay was performed after cells were cotransfected with IRF-3–Luc or NF-κB–Luc and other plasmids as indicated, and infected with SEV. As shown in Fig. 2a and b, the overexpression of PDCoV N protein blocked the SEV-induced promoter activities of IRF-3 (Fig. 2a) in a dose-dependent manner, but not NF-κB (Fig. 2b).
Fig. 2.
PDCoV N protein interrupts the activation of IRF3. a, b HEK-293T cells were co-transfected with IRF3–Luc (a), or NF-κB–Luc (b), together with the pRL–TK plasmid and increasing quantities (0, 0.2, 0.4, or 0.8 µg) of plasmid expressing PDCoV N protein. At 24 h post-transfection, cells were infected with SEV for 16 h and luciferase assays were performed. c, d HEK-293T cells were mock-transfected or transfected with an expression plasmid encoding HA-tagged N protein for 24 h and then mock-infected or infected with SEV for 8 h. c Cell lysates were collected for immunoblotting analysis with antibodies directed against phosphorylated IRF3 (Ser396), IRF3, HA, or β-actin. d Cells were fixed and permeated for immunofluorescence analysis to detect endogenous IRF3 (green) and N protein (red) with rabbit anti-IRF3 and mouse anti-HA antibodies. DAPI (blue) indicates the locations of the cell nuclei. Fluorescent images were acquired with a confocal laser scanning microscope (Olympus Fluoview ver. 3.1, Japan). Cells transfected with empty vector and mock-infected with SEV were used as the negative controls. e HEK-293T cells were cotransfected with IFN-β–Luc, pRL–TK plasmid, and pCAGGS-HA-PDCoV-N together with constructs expressing IRF3 or IRF3-5D. Luciferase assays were performed at 28 h post-transfection. ***P < 0.001; NS not significant
Phosphorylation and nuclear translocation are the hallmarks of IRF3 activation, which is essential for type I IFN transcription [31]. Because our initial results showed that PDCoV N protein blocked SEV-induced IRF3-dependent promoter activity, we further investigated the role of PDCoV N protein in the phosphorylation and nuclear translocation of IRF3. To this end, HEK-293T cells were transfected with pCAGGS-HA-PDCoV-N, and then infected with SEV. As expected, SEV infection markedly enhanced IRF3 phosphorylation compared with mock-treated cells. However, this increase was severely reduced in the N-protein-expressing cells (Fig. 2c). Consistent with this observation, nuclear translocation of IRF3 was also reduced by PDCoV N protein (Fig. 2d). Collectively, these results further supported the notion that PDCoV N protein is a potent inhibitor of type I IFN production by impeding the activation of IRF3.
To examine the role of PDCoV N protein downstream of IRF3 activation, we tested whether PDCoV N protein inhibits IFN-β promoter activation mediated by overexpressing IRF3 or IRF3-5D, a constitutively active mutant of IRF3. As shown in Fig. 2e, overexpression of IRF3 or IRF3-5D stimulated IFN-β promoter activation, but PDCoV N did not inhibit this activation, indicating that N protein inhibits the activation but not the function of IRF3. In addition, these results also suggested that the possible target(s) of PDCoV N protein to inhibit IFN-β production are likely located upstream of IRF3.
PDCoV N protein interacts with RIG-I
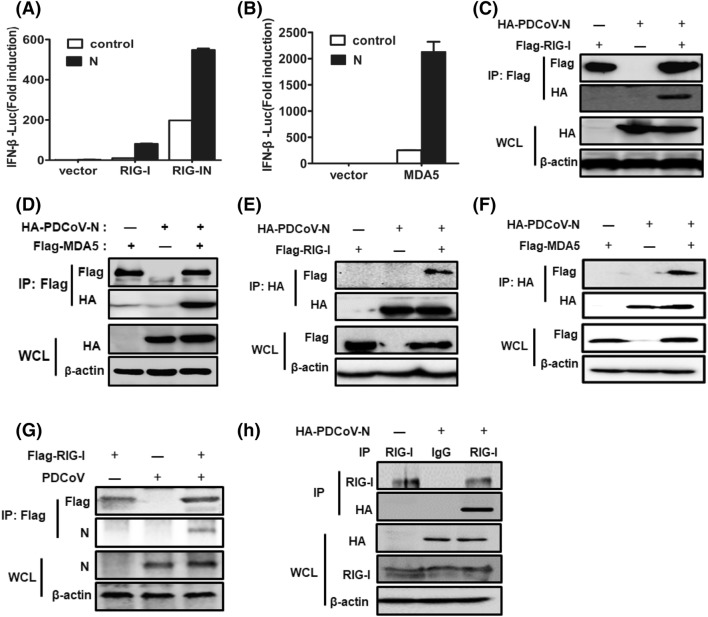
RIG-I and MDA5 are RNA sensors and can be activated by their respective RNA ligands to induce the downstream cascade pathway [32–36]. To identify the molecular target of PDCoV N protein in the RIG-I-like receptor (RLR) pathway, HEK-293T cells were co-transfected with pCAGGS-HA-PDCoV-N and luciferase reporter plasmids, IFN-β–Luc and pRL–TK, RIG/RIG-IN, or MDA5. Luciferase assays were performed at 28 h after co-transfection. The overexpression of RIG/RIG-IN and MDA5 dramatically stimulated IFN-β promoter activation. Interestingly, PDCoV N protein had no inhibitory effect on such activation (Fig. 3a and b). These results suggested that PDCoV N protein might target the protein(s) upstream of RIG-I/MDA5, or those associated with RIG-I/MDA5, and might even occur at RIG-I or MDA5.
Fig. 3.
PDCoV N protein interacts with RIG-I. a, b HEK-293T cells were co-transfected with IFN-β–Luc, pRL–TK plasmid, and pCAGGS-HA-PDCoV-N, together with constructs expressing RIG-I/RIG-IN or MDA-5. Luciferase assays were performed at 28 h after transfection. The results represent the means and standard deviations of three independent experiments. c–f HEK-293T cells were co-transfected with expression plasmids encoding HA-tagged N protein and Flag-tagged RIG-I or MDA5. The cells were lysed at 28 h post-transfection and subjected to immunoprecipitation with anti-Flag (c, d) or anti-HA (e, f) antibodies. The whole-cell lysates (WCL) and immunoprecipitate (IP) complexes were analyzed by immunoblotting with anti-Flag, anti-HA, or anti-β-actin antibodies. g LLC-PK1 cells transfected RIG-I expression plasmids were infected with PDCoV. Cells were then lysed for immunoprecipitation with anti-Flag antibodies. Lysates and immunoprecipitates were subjected to immunoblotting with anti-PDCoV N, anti-Flag, or anti-β-actin antibodies. h HEK-293T cells were transfected with HA-tagged N protein only, then cells were lysed and immunoprecipitated with endogenous anti-RIG-I antibodies. Lysates and IPs were analyzed by immunoblotting with anti-RIG-I, anti-HA, or anti-β-actin antibodies
To further verify the hypothesis that the inhibition of IFN-β production by N protein occurs at the RIG-I/MDA5-dsRNA recognition step, potential interactions between N protein and RIG-I/MDA5 were investigated. We co-transfected HA-tagged PDCoV N protein and Flag-tagged RIG-I or MDA5 into HEK-293T cells to perform co-immunoprecipitations (co-IPs) with anti-Flag antibodies. As shown in Fig. 3c and d, HA-N protein was detected in Flag-tagged RIG-I or MDA5 precipitates, suggesting that PDCoV N protein interacts with RIG-I and MDA5. The interaction between N protein and RIG-I or MDA5 was reconfirmed by using a reverse co-IP assay with anti-HA antibody immunoprecipitating lysates. And as shown in Fig. 3e and f, RIG-I and MDA5 were also pulled down by HA-N protein.
To further investigate whether the interactions between N protein and RIG-I/MDA5 occur in PDCoV-infected cells, LLC-PK1 cells were transfected with the Flag-tagged RIG-I or MDA5 expression constructs, followed by PDCoV infection. Co-IPs with anti-Flag antibodies were performed and the N protein was detected with monoclonal antibodies against PDCoV N protein. As shown in Fig. 3g, the N protein could be readily immunoprecipitated by Flag-RIG-I. When LLC-PK1 cells were transfected with pCAGGS-HA-PDCoV-N and then immunoprecipitated by antibody against RIG-I, the N protein could also be readily pulled down by endogenous RIG-I (Fig. 3h). It should be noted that occasionally, only weak N-MDA5 interactions could be detected in the context of PDCoV infection or pulled down by endogenous MDA5 (data not shown). Thus, we focused on the RIG-I-counteracting mechanism in the subsequent studies.
PDCoV N protein interacts with the helicase domain and the C-terminal domain of RIG-I
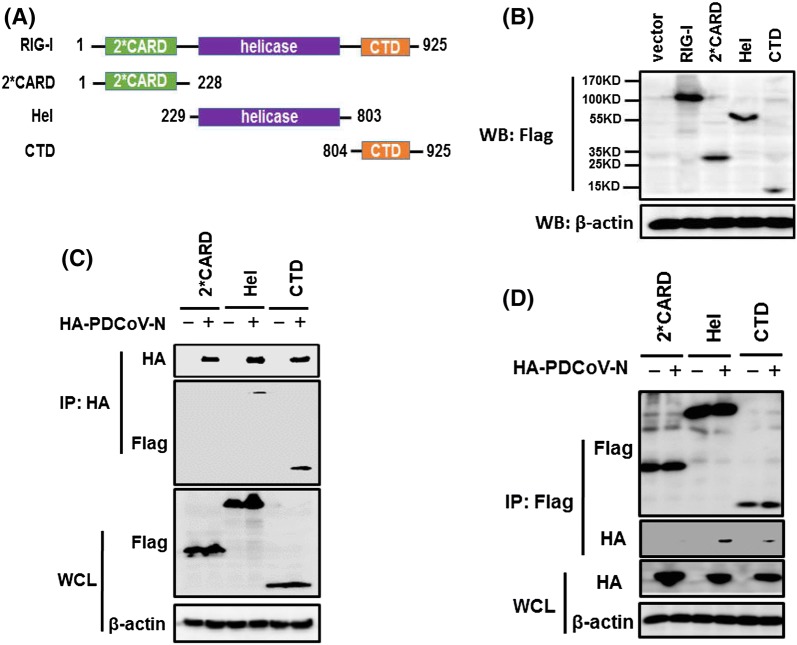
Structurally, RIG-I comprises three functional domains: two N-terminal caspase activation and recruitment domains (2*CARD), a helicase domain and a CTD (Fig. 4a) [37–39]. The corresponding unbiased map was used to build the 2*CARD, helicase domain and CTD truncated structures, and these mutants were confirmed as normally expressed in transfected HEK-293T cells (Fig. 4b). To identify the potential domain(s) of RIG-I targeted by PDCoV N protein, we co-transfected vectors expressing HA-tagged PDCoV N protein and the truncated RIG-I containing only the 2*CARD, helicase domain or CTD into HEK-293T cells. Immunoprecipitation analysis with anti-HA (Fig. 4c) or -Flag (Fig. 4d) antibodies showed that the CTD and helicase domain, but not 2*CARD, co-immunoprecipitated with PDCoV N protein. These findings suggested that the helicase domain and the CTD were responsible for the interaction with PDCoV N protein.
Fig. 4.
PDCoV N protein interacts with the helicase domain and the CTD of RIG-I. a Architectures of wild type RIG-I and deletion constructs. 2*CARD, two tandem repeat caspase activation recruitment domains (green); Hel, conserved RecA-like domains in the DExD/H helicase family (purple); CTD, C-terminal domain (orange). Numbers indicate the amino acid residue number. b HEK-293T cells were transfected with mock vectors, wild type RIG-I or RIG-I deletion constructs. At 28 h post-transfection, immunoblots were performed with anti-Flag or anti-β-actin antibodies. (c and d) HEK-293T cells were co-transfected with HA-tagged PDCoV N protein and Flag-tagged 2*CARD, Hel or CTD constructs. At 28 h post-transfection, immunoprecipitation was performed with anti-HA (c) or anti-Flag antibodies (d). WCL and IP complexes were analyzed by immunoblotting with anti-Flag, or anti-HA, or anti-β-actin antibodies
PDCoV N protein interferes with dsRNA and PACT binding to RIG-I
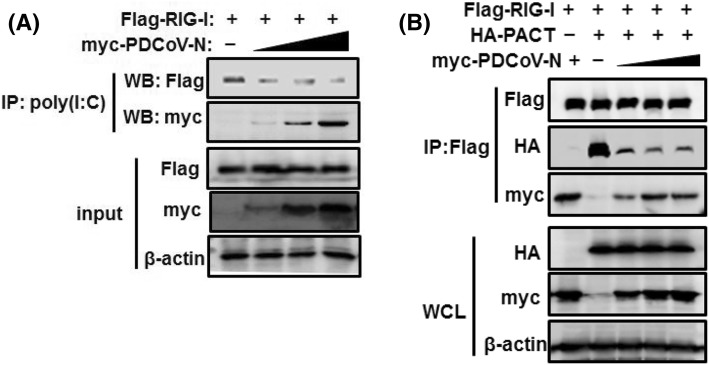
Previous studies have shown that the helicase domain and CTD of RIG-I can bind to dsRNA [36, 40]. Interactions of PDCoV N protein with the helicase domain and CTD may affect the binding of RIG-I to dsRNA. Further, we tested the influence of PDCoV N protein on RIG-I recognition of dsRNA. Increasing amounts of pCAGGS-Myc-PDCoV-N expression vector along with constant PEF-Bos-RIG-I were co-transfected into HEK293T cells. Cell lysates were incubated with poly(I:C) to detect dsRNA binding ability. As shown in Fig. 5a, RIG-I was strongly pulled down by poly(I:C), but this was decreased in the presence of PDCoV N protein. The results suggested that N protein attenuated RIG-I recognition of RNA.
Fig. 5.
PDCoV N protein impairs poly(I:C) and PACT binding to RIG-I. a HEK-293T cells were co-transfected with constant Flag-tagged RIG-I and increasing Myc-tagged PDCoV N expressing plasmids. At 28 h post-transfection, cell lysates were incubated with poly(I:C)-coated agarose at 4 °C for 4 h. Lysates and precipitates were subjected to immunoblot analysis using anti-Flag, anti-Myc or anti-β-actin antibodies. b HEK-293T cells were co-transfected with HA-tagged PACT, Flag-tagged RIG-I vectors and pCAGGS-Myc-PDCoV-N. Transfected HA-tagged PACT, Flag-tagged RIG-I vectors were kept constant, and pCAGGS-Myc-PDCoV-N constructs were increased. At 28 h post-transfection, the cells were harvested for immunoprecipitation with anti-Flag antibodies and immunoblotted with antibodies against HA, Flag, Myc or β-actin
The CTD of RIG-I also binds to PACT, a cellular dsRNA-binding protein that can bind to RIG-I and MDA5 to activate IFN production [40]. Then, we assessed the ability of PACT to associate with RIG-I when increasing concentrations of pCAGGS-Myc-PDCoV-N expression vectors were transfected. Immunoprecipitation was performed with anti-Flag antibody after co-transfection with Flag-tagged RIG-I and HA-tagged PACT, together with increasing Myc-tagged N plasmid. Precipitates and lysates were analyzed by immunoblotting with anti-Myc, -Flag or -HA antibodies. As shown in Fig. 5b, RIG-I efficiently pulled down PACT, however, the amount of PACT in RIG-I precipitates was dramatically reduced as PDCoV N protein expressed. These results indicated that PDCoV N proteins interferes with RIG-I interaction with PACT.
Discussion
CoVs are positive-sense RNA viruses that generate dsRNA intermediates during replication, which can trigger the host innate immune defense [41]. However, cells infected with PDCoV fail to induce type I IFN [42]. Previous studies have shown that nsp5 of PDCoV cleaves NF-κB essential modulator (NEMO) and signal transducer and activator of human transcription 2 (STAT2) to suppress type I IFN production and its associated signaling pathway, respectively [43, 44]. Recently, PDCoV accessory protein 6 (NS6) was also identified as an IFN-β antagonist through blockade of RIG-I/MDA5 binding to dsRNA [45]. In this study, we have shown that N protein acts as another IFN antagonist by at least two strategies: (i) interfering with the interaction between RIG-I and PACT; (ii) attenuating the binding of dsRNA to RIG-I.
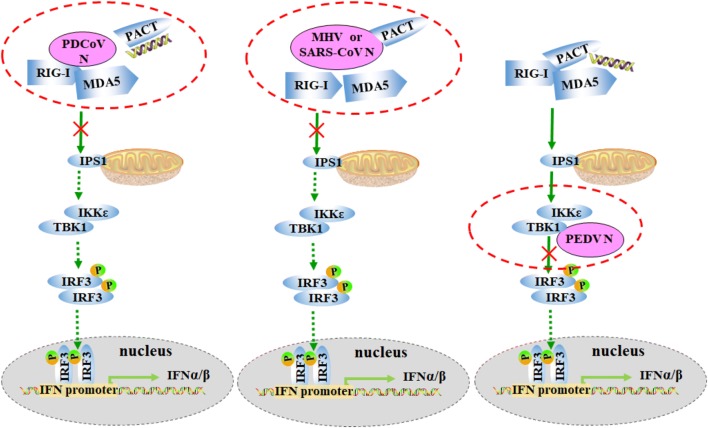
CoV N proteins share a similar overall topology structure and a highly conserved function of packaging the viral genome with structural proteins to form ribonucleoprotein complexes for viral assembly [46]. Nevertheless, they also manifested inhibitory trends in innate immune mediation and the strategies they exploit are complicated. The N protein from PEDV (alphacoronavirus) used a totally distinct mechanism from other CoVs, targeting TBK1 and impairing TBK1 recruitment and phosphorylation of IRF3 [23]. N proteins from MHV and SARS-CoV (betacoronavirus) were shown to target PACT to inhibit IFN-β synthesis [24], but the N protein of MHV also inhibited RNaseL-mediated host translational shut-off [47], which has not yet been shown for SARS-CoV N protein. The MERS-CoV N protein was previously demonstrated to be unable to suppress IFN production [48] but recently, SARS-CoV and MERS-CoV N proteins were discrepantly reported to interact with TRIM25 and interfere with TRIM25-mediated RIG-I ubiquitination [25]. The IFN-β inhibitory tactics used by PDCoV (deltacoronavirus) N protein showed some similarity to MHV and SARS-CoV N proteins, which function upstream of RLR molecules. However, distinct from other CoV N proteins, PDCoV N protein interacts with RIG-I, and our results clearly show that this interaction leads to the blockade of RIG-I binding to its agonists dsRNA and PACT, as shown in Fig. 6.
Fig. 6.
Schematic diagram of coronavirus N proteins antagonizing IFN-β production. RIG-I and MDA5 sense RNA PAMPs or/and bind to PACT, which form the complex, and activate the downstream IKK/TANK-binding kinase 1 (TBK-1) and the IKKε through IPS-1, leading to activation of interferon regulatory factor 3 (IRF3). The activated IRF3 translocates into the nucleus and activates the transcription of type I interferons. The molecules in RIG-I like receptor (RLR) cascades were blue marked. The targets and tactics of N proteins (pink marked) from PDCoV, MHV, SARS-CoV and PEDV bind to or developed were circled by dotted red line
The activation of RIG-I is a multi-step process and each domain of RIG-I functions uniquely in autorepression and activation. Original CARDs and Hel2i interactions hinder dsRNA binding and inactivate RIG-I [39, 49, 50]. Binding of cytoplasmic virus-specific RNA structures to the CTD leads to a conformational switch of RIG-I, which releases the autorepressed CARDs and exposes the helicase domain for ATP binding [32, 50–52]. The process enables RIG-I to translocate along the dsRNA, and further promotes the oligomerization of RIG-I CARDs, assembling RIG-I into a filamentous architecture which facilitates CARD–CARD interactions with mitochondrial MAVS for signal transduction [53, 54]. In addition, ubiquitination, phosphorylation and acetylation modifications are also involved in these processes [52, 55]. Because RIG-I/MDA5 are important cytoplasmic PRRs that recognize pathogen-associated molecular patterns derived from viruses and initiate the type I IFN response against infection, it is not surprising that many viral proteins target these two molecules to disrupt IFN signaling. At least five mechanisms used by viral proteins have been reported to disable RIG-I-mediated signaling: (i) preventing RNA recognition by RIG-I, for example, the Vpg of poliovirus is linked to the genomic RNA to cap the 5′-end from exposure to RIG-I [56]; (ii) interfering with PACT-RIG-I complex conformation. MHV and SARS-CoV N proteins interact with PACT to disrupted PACT-mediated RIG-I activation for IFN production [24]; (iii) directly cleaving or degrading RIG-I, for example, RIG-I has been reported to be cleaved by the proteinase 3Cpro during infections with picornavirus, coxsackievirus B3, and enterovirus 71 [57–59], and the encephalomyocarditis virus directs both caspase- and proteasome-dependent degradation of RIG-I [60]; (iv) inhibiting RIG-I–MAVS signal transduction, for example, the PB1-F2 of influenza A virus (IAV) has been described to decrease the mitochondrial membrane potential, resulting in the acceleration of mitochondrial fragmentation, thereby inhibiting RIG-I–MAVS signaling [61–63]; (v) affecting the post-translation modification of RIG-I. IAV NS1 has been reported to bind to TRIM25 and Riplet, preventing RIG-I K63-ubiquitination, and therefore, downstream binding to MAVS [64, 65]. The observation in this study that PDCoV N protein did not subvert RIG-I/RIG-IN activation or expression suggested that PDCoV N protein does not counteract RIG-I signaling transduction, or cleave or degrade RIG-I. Our investigation at least supports the possibility that PDCoV N protein hides RIG-I from associating with dsRNA or PACT.
RIG-I and MDA5 sense complementary sets of viral RNA ligands: RIG-I recognizes short dsRNA and 5′-phosphorylated blunt ends of viral genomic dsRNA; MDA5 binds internally to long dsRNA with no end specificity [66–68]. However, they both are important for coronavirus recognition. Previous studies have shown that MHV can be recognized by both RIG-I and MDA5 in mouse oligodendrocytic N20.1 cells [69]. In our previous study, we also found that both RIG-I and MDA5 play roles in the induction of innate immune responses by TGEV, another swine enteropathogenic coronavirus [70]. MDA5 shows a similar structure to RIG-I, comprising CARDs with death domain folds, a DExD/H-box helicase (consisting of two RecA-like helicase domains, Hel1 and Hel2, and an insert domain, Hel2i), and a CTD [71]. Considering the high similarity of structure, PDCoV N protein may also target MDA5. However, the structure and function of the MDA5 CTD is different from RIG-I, as MDA5 does not contain a conserved internal repressor domain and the MDA5 CTD is required for cooperative filament assembly but not for RNA binding [71, 72]. This may contribute to the N–MDA5 interaction in a weak or dynamic way, which can only and occasionally be proven in PDCoV-infected cells.
Acknowledgment
We thank Dr. Zhigao Bu for providing VSV-GFP recombinant virus. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31730095, 31860704), the National Key R&D Plan of China (Grant No. 2016YFD0500103), the Open Research Fund Program of the State Key Laboratory of Agricultural Microbiology (Grant No. AMLKF201805) and Jiangxi Province Project (Grant No. GJJ170261).
Author contributions
ZD, LF, GP, and SX designed the experiments; CJ, PF, QP, JR and MW carried out the experiments; CJ, DW and GP analyzed the data; JC wrote the first draft; JC, ZD, SX and LF reviewed and edited the manuscript.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Research involving human and animal participants
This article did not contain any study with human participants or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, Bai R, Teng JLL, Tsang CCC, Wang M, Zheng B, Chan K, Yuen K. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J (2014) Full-Length Genome Sequence of Porcine Deltacoronavirus Strain USA/IA/2014/8734. Genome Announc 2 [DOI] [PMC free article] [PubMed]
- 6.Marthaler D, Jiang Y, Collins J, Rossow K (2014) Complete Genome Sequence of Strain SDCV/USA/Illinois121/2014, a Porcine Deltacoronavirus from the United States. Genome Announc 2 [DOI] [PMC free article] [PubMed]
- 7.Lee S, Lee C (2014) Complete Genome Characterization of Korean Porcine Deltacoronavirus Strain KOR/KNU14-04/2014. Genome Announc 2 [DOI] [PMC free article] [PubMed]
- 8.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in china: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madapong A, Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Srijangwad A, Tripipat T, Wegner M, Nilubol D (2016) Complete Genome Sequence of Porcine Deltacoronavirus Isolated in Thailand in 2015. Genome Announc 4 [DOI] [PMC free article] [PubMed]
- 10.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. Different Lineage of Porcine Deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis. 2017;64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung K, Hu H, Saif LJ. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung K, Hu H, Saif LJ. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C, Chen P, Yeh SH. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo L, Koetzner CA, Masters PS. A key role for the carboxy-terminal tail of the murine coronavirus nucleocapsid protein in coordination of genome packaging. Virology. 2016;494:100–107. doi: 10.1016/j.virol.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C, Chen C, Chiang M, Hsu Y, Huang T. Transient oligomerization of the SARS-CoV N protein–implication for virus ribonucleoprotein packaging. PLoS ONE. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Qu J. Differential characteristics of the early stage of lung inflammation induced by SARS-CoV Nucleocapsid protein related to age in the mouse. Inflamm Res. 2009;58:312–320. doi: 10.1007/s00011-009-8062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Zhang H, Zhang Q, Huang Y, Dong J, Liang Y, Liu H, Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L, Wang H, Ji Y, Yang J, Xu S, Huang X, Wang Z, Qin L, Tien P, Zhou X, Guo D, Chen Y. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J Virol. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, Xu X, Tong D. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun. 2014;445:497–503. doi: 10.1016/j.bbrc.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Z, Fang L, Yuan S, Zhao L, Wang X, Long S, Wang M, Wang D, Foda MF, Xiao S. The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-beta antagonizing mechanism: attenuation of PACT-mediated RIG-I/MDA5 activation. Oncotarget. 2017;8:49655–49670. doi: 10.18632/oncotarget.17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol. 2017;91:e02143. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. Porcine deltacoronavirus in Mainland China. Emerg Infect Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siu KL, Yeung ML, Kok KH, Yuen K, Kew C, Lui P, Chan C, Tse H, Woo PC, Yuen K, Jin DY. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 30.Sharma S, tenOever BR, Grandvaux N, Zhou G, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 31.Ramos HJ, Gale MJ. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 34.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 35.Wu B, Hur S. How RIG-I like receptors activate MAVS. Curr Opin Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C, Ranjith-Kumar CT, Hao L, Kao C, Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo D, Kohlway A, Pyle AM. Duplex RNA activated ATPases (DRAs): Platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–120. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou T, Hu F, Herr AB, Strong RK, Kao C, Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohlway A, Luo D, Rawling DC, Ding S, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kok KH, Lui P, Ng MH, Siu KL, Au SW, Jin DY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9:299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Sola I, Almazan F, Zuniga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Fang L, Dong N, Fang P, Ding Z, Wang D, Chen H, Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X, Fang L, Wang D, Yang Y, Chen J, Ye X, Foda MF, Xiao S. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology. 2017;502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, Lv M, Ye X, Peng G, Fang L, Xiao S. Porcine Deltacoronavirus nsp5 Antagonizes Type I Interferon Signaling by Cleaving STAT2. J Virol. 2017;91:e00003–e00017. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang P, Fang L, Ren J, Hong Y, Liu X, Zhao Y, Wang D, Peng G, Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J Virol. 2018;92:e00712–e00718. doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding B, Qin Y, Chen M. Nucleocapsid proteins: roles beyond viral RNA packaging. Wiley Interdiscip Rev RNA. 2016;7:213–226. doi: 10.1002/wrna.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 50.Luo D, Ding S, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang F, Ramanathan A, Miller MT, Tang G, Gale MJ, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen Z. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 55.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 56.Lee YF, Nomoto A, Detjen BM, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, Emdad L, Bhoopathi P, Fisher PB, Lloyd RE, van Kuppeveld FJ. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology. 2009;393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 61.Varga ZT, Grant A, Manicassamy B, Palese P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol. 2012;86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- 64.Gack MU, Albrecht RA, Urano T, Inn KS, Huang I, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajsbaum R, Albrecht RA, Wang M, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 68.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Liu Y, Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding Z, An K, Xie L, Wu W, Zhang R, Wang D, Fang Y, Chen H, Xiao S, Fang L. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology. 2017;507:170–178. doi: 10.1016/j.virol.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reikine S, Nguyen JB, Modis Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale MJ. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]