Abstract

DNA damage activates checkpoints to arrest cell cycle progression in S and G2 phases, thereby providing time for repair and recovery. The combination of DNA-damaging agents and inhibitors of CHK1 (CHK1i) is an emerging strategy for sensitizing cancer cells. CHK1i induce replication on damaged DNA and mitosis before repair is complete, and this occurs in a majority of cell lines. However, ∼15% of cancer cell lines are hypersensitive to single-agent CHK1i. As both abrogation of S phase arrest and single-agent activity depend on CDK2, this study resolved how activation of CDK2 can be essential for both replication and cytotoxicity. S phase arrest was induced with the topoisomerase I inhibitor SN38; the addition of CHK1i rapidly activated CDK2, inducing S phase progression that was inhibited by the CDK2 inhibitor CVT-313. In contrast, DNA damage and cytotoxicity induced by single-agent CHK1i in hypersensitive cell lines were also inhibited by CVT-313 but at 20-fold lower concentrations. The differential sensitivity to CVT-313 is explained by different activity thresholds required for phosphorylation of CDK2 substrates. While the critical CDK2 substrates are not yet defined, we conclude that hypersensitivity to single-agent CHK1i depends on phosphorylation of substrates that require high CDK2 activity levels. Surprisingly, CHK1i did not increase SN38-mediated cytotoxicity. In contrast, while inhibition of WEE1 also abrogated S phase arrest, it more directly activated CDK1, induced premature mitosis, and enhanced cytotoxicity. Hence, while high activity of CDK2 is critical for cytotoxicity of single-agent CHK1i, CDK1 is additionally required for sensitivity to the drug combination.
Keywords: CHK1, WEE1, CDK2, DNA damage, topoisomerase I inhibition, S phase arrest, checkpoint abrogation
Introduction
Many anticancer agents elicit cytotoxicity as a consequence of inducing damage to DNA. This DNA damage activates cell cycle checkpoints to arrest cell cycle progression in S and G2 phases, thereby permitting time for repair. Inhibitors of checkpoint kinase 1 (CHK1i) have been developed as potential therapeutic agents to combine with DNA-damaging drugs because they can induce replication on damaged DNA and mitosis before replication and repair are complete.1 In contrast, cells arrested in S phase by gemcitabine, a ribonucleotide reductase inhibitor, cannot progress through S phase upon addition of CHK1i because they lack dNTPs. In these cells, CHK1i trigger dormant origin firing leading to extensive single-strand DNA that is subject to nuclease attack, resulting in replication catastrophe.2,3
While CHK1i-mediated replication and mitotic catastrophe appear to occur in most, if not all, cell lines, we have demonstrated that ∼15% of cell lines are hypersensitive to CHK1i as a single agent.4 However, whether the cytotoxic mechanism for CHK1i as a single agent is similar to its action in combination with DNA-damaging drugs remains unclear. Here, we have further analyzed the mechanism of CHK1i-mediated sensitization to the topoisomerase I inhibitor SN38 and compared it to the mechanism of action of CHK1i as a single agent.
Normal cell cycle progression is regulated by cyclin-dependent kinases (CDK). CDK2 is required for S phase entry, while CDK1 is required for mitosis.5 Entry into the S phase also requires the activity of DBF4-dependent CDC7 kinase.6 Formation of the pre-replication complex requires loading of the minichromosome maintenance protein MCM2–7 helicase complex onto chromatin; CDC7 then phosphorylates MCM2–7 to trigger origin firing.7,8 Concurrently, CDK2 phosphorylates treslin, which recruits the helicase cofactor, CDC45, to fully activate the helicase and recruit additional regulatory proteins.9 Many more pre-replication complexes are formed than required for normal DNA synthesis, but these dormant origins can fire to circumvent damage-induced blocks to DNA synthesis.
CDK2 is generally believed to be required for both initiation and progression through S phase, as CDC45 chromatin loading is required for firing of both early and late origins of replication.10 Furthermore, active CDK2 accumulates as the S phase progresses.11 Therefore, it is surprising that inhibition of CDK2 arrests asynchronous cells in the G1 phase but not in the S phase.4,12 Furthermore, we recently demonstrated that cells poorly tolerate CDK2 activity in the S phase.4 Specifically, incubation with the CHK1i, MK-8776, as a single agent can induce extensive DNA breaks in S phase cells in a CDK2/cyclin A-dependent manner. The DNA breaks were also suppressed by inhibition of either MRE11 or MUS81,13 but the mechanism by which CDK2 activates these nucleases remains to be established. Most cell lines are resistant to CHK1i because they are able to prevent CDK2 activation in the S phase cells. This has been attributed to a failure of CHK1i to activate CDC25A, the phosphatase that dephosphorylates and activates CDK2. In contrast, most cell lines are sensitive to the WEE1 inhibitor (WEE1i) AZD1775, which directly activates CDK1 and CDK2, yet the DNA breaks again occur in the S phase and are dependent on CDK2.4
Topoisomerase I inhibitors such as SN38 are frequently used to induce S phase arrest. Arrest is mediated by activation of CHK1 that directly suppresses CDC25A phosphatase and as a consequence inhibits CDK2. Inhibition of CHK1 causes re-accumulation of CDC25A and S phase progression, and it has logically been presumed that this is dependent on CDK2 activation.14−17 This would appear to be inconsistent with the observation described above that cells do not tolerate CDK2 activity in the S phase. Surprisingly, we demonstrate here that the concentrations of CDK2i that prevent CHK1i single-agent cytotoxicity fail to prevent CHK1i-mediated abrogation of SN38-mediated S phase arrest.
While trying to resolve this disconnect, we found that much higher concentrations of CDK2i prevent CDC45 loading and S phase progression of damaged cells. This unexpected observation is consistent with recent reports in yeast that the efficacy of CDK inhibitors can vary depending on the particular substrate.18,19 Considering that the sensitivity to CHK1i in damaged cells is inhibited only at high concentrations of CDK2i while the single-agent activity of CHK1i is inhibited at much lower concentrations, we conclude that different substrates of CDK2 are involved in these disparate mechanisms of action. This study also compares the efficacy of CHK1i and WEE1i to promote premature mitotic entry and enhance cytotoxicity in SN38-damaged cells. We demonstrate that the combination with WEE1i is more cytotoxic than CHK1i presumably as it more directly activates CDK1.
Results
Abrogation of S Phase Arrest by the CHK1i MK-8776 or the WEE1i AZD1775 Is Insensitive to Inhibition of CDK1/2
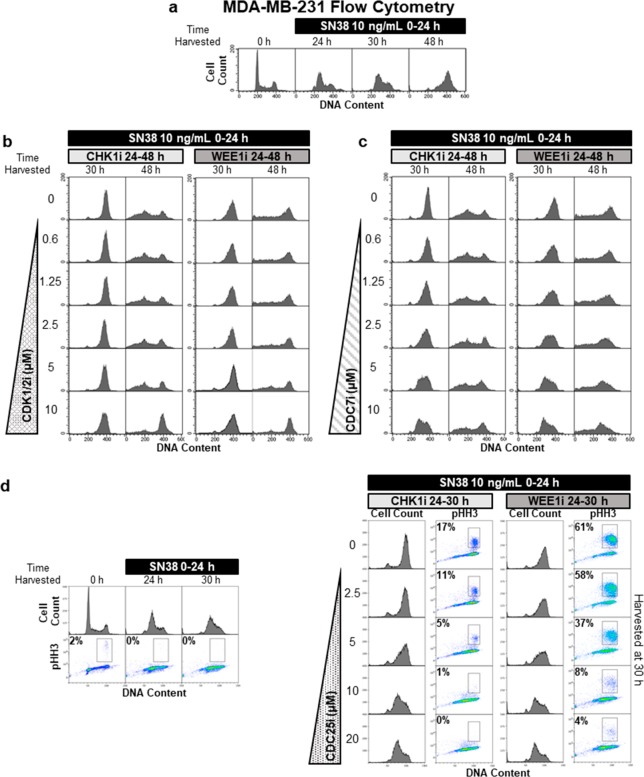
Initial experiments were designed to assess the potential role of CDK1 and CDK2 in abrogation of S and G2 phase arrest induced by SN38. These experiments were performed with MDA-MB-231 cells because of our extensive prior analysis of SN38 in these cells14,20 and because they are resistant to MK-8776 as a single agent at the concentrations used.4 Cells were incubated for 24 h with a concentration of SN38 that induced arrest in the mid S phase (Figure 1a). Upon removal of SN38, cells slowly progressed through the S phase and were arrested in the G2 phase within the following 24 h (48 h time point). Inhibiting CHK1 following SN38 removal (24 h) dramatically accelerated S phase progression (Figure 1b); the majority of cells reached the G2 phase by 30 h. Additionally, many cells exhibited sub-G1 phase DNA content by 48 h, suggesting cell death following aberrant mitosis.
Figure 1.
Abrogation of S phase arrest by CHK1i or WEE1i is insensitive to inhibition of CDK1/2 but requires CDC7 and CDC25 activity. (a) MDA-MB-231 cells were incubated with SN38 for 24 h and harvested as indicated. Cells were stained for DNA with propidium iodide and analyzed by flow cytometry. (b) Following removal of SN38 at 24 h, cells were incubated with 1 μM MK-8776 (CHK1i) or 1 μM AZD1775 (WEE1i) concurrently with 0–10 μM CVT-313 (CDK1/2i). (c) Same as panel b, except 0–10 μM XL413 (CDC7i) was added concurrently with CHKi or WEE1i. (d) MDA-MB-231 cells were incubated with SN38 for 0–24 h, and then CHK1i or WEE1i were added from 24 to 30 h, concurrently with 0–20 μM NSC663284 (CDC25i). Cells were immunostained with a fluorescent anti-pHH3 antibody, and DNA was stained with propidium iodide and then analyzed by flow cytometry. The gates and percentages shown represent the proportion of cells positive for pHH3.
To assess the role of CDK1 and/or CDK2 in abrogation of S phase arrest, we used CVT-313, which is the most selective inhibitor of CDK2 available but also inhibits CDK1 at higher concentrations.21 We previously demonstrated that 2 μM CVT-313 prevented DNA breaks as assessed by the comet assay, and γH2AX induced by single-agent MK-8776 treatment, while 5 μM CVT-313 prevented cells from entering the S phase, both events considered to be a consequence of CDK2 activity.4 Following SN38, administration of 10 μM CVT-313 did not prevent CHK1i-mediated S phase progression but did prevent lethal mitotic entry as predicted from its ability to inhibit CDK1 (Figure 1b). Hence, it appeared that S phase progression occurred independently of CDK1 and CDK2.
The WEE1 inhibitor, AZD1775, directly activates CDK1 and CDK2 by preventing phosphorylation of tyrosine 15.4 We hypothesized that WEE1i would also abrogate S and G2 phase arrest induced by SN38. WEE1i was effective at abrogating S phase arrest and driving cells through the G2/M phase (Figure 1b). It is worth emphasizing that WEE1i did not induce a direct S to M phase transition as has been observed in a few cell lines;22,23 rather, the cells progressed to the G2 phase before undergoing mitosis. CVT-313 at 10 μM failed to prevent abrogation of S phase arrest but inhibited mitosis. These results again suggested that CDK2 is not required for S phase progression of damaged cells. It was particularly unexpected that the bypass of S phase arrest by AZD1775 would be insensitive to inhibition of CDK1/2 as these are the only known substrates of WEE1.
CDC7 Is Required for Abrogation of S Phase Arrest by CHK1i and WEE1i
S phase entry and progression also require CDC7-mediated phosphorylation of the MCM2–7 complex. Considering the insensitivity of S phase progression of damaged cells to CDK1/2i, we investigated the role of CDC7. The CDC7 inhibitor XL413 prevented S phase progression induced by both CHK1i and WEE1i (Figure 1c). Interestingly, XL413 did little to prevent the appearance of sub-G1 cells at 48 h. Other experiments presented below suggest this may be due to the cells prematurely entering mitosis from the S phase.
CDC25 Is Required for S Phase Progression and Mitotic Entry
The results presented above suggested that abrogation of S phase arrest did not depend on the canonical CHK1–CDC25–CDK2 signaling axis. To further explore regulation of S phase abrogation, we tested the involvement of CDC25 using the pan CDC25 inhibitor, NSC663284.24 Concentrations of NSC663284 reported to inhibit CDC25 prevented abrogation of arrest by CHK1i (Figure 1d). We also assessed the phosphorylation of serine 10 on histone H3 (pHH3), a common marker of mitosis. CHK1i induced pHH3 in 17% of cells but only after they progressed to the G2/M phase (Figure 1d). Furthermore, NSC663284 clearly inhibited pHH3 at concentrations that also retarded S phase progression.
The addition of WEE1i to SN38-arrested cells induced pHH3 in a much greater proportion of the cells, but again only after they had progressed to the late S or G2/M phase (Figure 1d). NSC663284 also prevented abrogation of arrest, and the appearance of pHH3, by WEE1i. This can presumably be explained because CDC25 phosphatase is still required to remove any existing phosphates to activate CDK1/2 even when WEE1 is inhibited.
Analysis of CDK1/2 and CDC7 Substrates
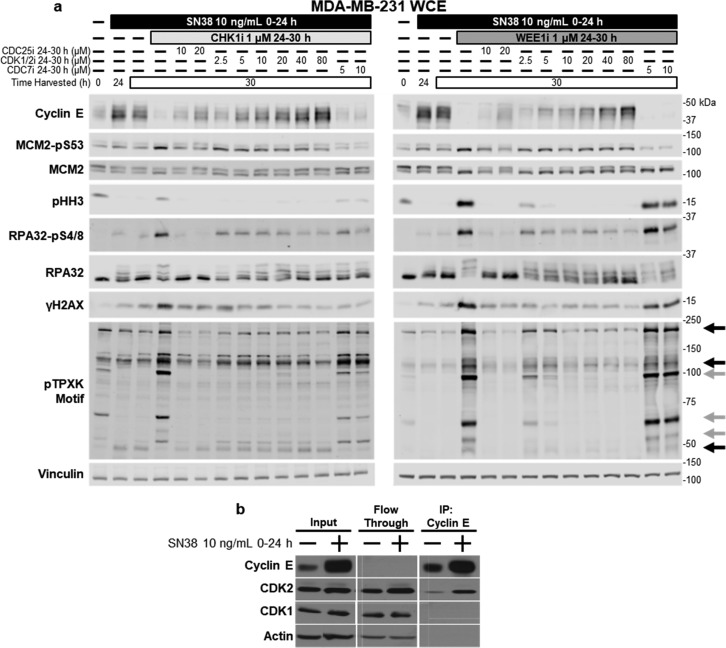
To further dissect the roles of CDK1/2 and CDC25 in S phase progression, we investigated downstream targets. The level of cyclin E protein is inversely related to the activity of CDK2, because CDK2 phosphorylates cyclin E thereby targeting it for degradation.25−27 Incubation of MDA-MB-231 cells with SN38 for 24 h caused dramatic accumulation of cyclin E, consistent with inhibition of CDK2 (Figure 2a). Following removal of SN38, cyclin E levels remained high for an additional 6 h. CHK1i significantly decreased the level of cyclin E protein during this 6 h period, consistent with activation of CDK2. However, in the initial experiments, we found that concentrations of CVT-313 of ≤10 μM only partially rescued cyclin E; similar results were obtained with WEE1i. Increasing the CVT-313 concentration to 80 μM completely prevented degradation of cyclin E protein following CHK1i and WEE1i (Figure 2a). This suggested that high concentrations of CVT-313 are required to fully inhibit CDK2. High concentrations of CVT-313 also prevented S phase progression as will be discussed below. Additionally, CDC25i partially prevented degradation of cyclin E, consistent with preventing activation of CDK2 (Figure 2a).
Figure 2.
CDC25i and CDK1/2i, but not CDC7i, prevent pHH3 and γH2AX from SN38 with CHK1i or WEE1i. (a) MDA-MB-231 cells were incubated with SN38 for 24 h and then CHK1i or WEE1i from 24 to 30 h. Additionally, CDC25i, CDK1/2i, or CDC7i was administered concurrently with CHK1i or WEE1i as indicated. Whole cell extracts were probed for the indicated proteins by Western blotting. Black arrows indicate bands relatively resistant to CVT-313, and gray arrows indicate bands very sensitive to CVT-313; these arrows highlight the same bands noted in Figure 4b. (b) MDA-MB-231 cells were incubated with SN38 for 24 h. Cyclin E protein was immunoprecipitated, and the indicated proteins were probed by Western blotting.
Considering the large increase in the level of cyclin E following SN38 treatment, we immunoprecipitated cyclin E and assessed its binding partners. It has been reported that cyclin E can bind CDK1 in cells lacking CDK2.28 We questioned whether the dramatic increase in the level of cyclin E might allow binding to CDK1, and hence, CDK1 could contribute to the turnover of cyclin E. However, only CDK2 was detected in the immunoprecipitates (Figure 2b). The amount of CDK2 bound to cyclin E was increased compared to that in untreated cells, but most of the CDK2 remained unbound to cyclin E. Some remaining CDK2 was likely bound to cyclin A, although the majority usually remains unbound to either cyclin.29 Hence, this is consistent with the involvement of CDK2 rather than CDK1 in the degradation of cyclin E.
We next assessed phosphorylation of MCM2 at serine 53, a target of CDC7 required for origin firing.7,8 Two phosphorylated bands were observed (Figure 2a), but the more rapidly migrating band is generally considered the active form.30 This phosphorylated band was also observed as the faster-migrating band when probing for total MCM2 (Figure 2a). Incubation with SN38 slightly increased the intensities of both of the phosphorylated bands, demonstrating that CDC7 remains active in arrested cells as previously reported.31,32 The addition of either CHK1i or WEE1i shifted the majority of MCM2 to the rapidly migrating, phosphorylated form (Figure 2a). Concurrent incubation with CDK1/2i had little effect on the intensity of the bands. Incubation with CDC7i caused a major decrease in the levels of both phospho forms, as expected. Additionally, CDC7i did not prevent cyclin E degradation despite preventing abrogation of S phase arrest.
Analysis of pHH3 and γH2AX Induced by SN38 with CHK1i or WEE1i
Concurrently, we assessed other markers that might predict the outcome of these drug combinations. The level of phosphorylation of histone H3 (pHH3), a marker of mitosis, was slightly increased by CHK1i but dramatically increased by WEE1i (Figure 2a), consistent with flow cytometry results (Figures 1d and 3). Both CDC25i and CDK1/2i prevented pHH3, which in the case of CVT-313 occurred at low concentrations (≤2.5 μM) (Figures 1d, 2a, and 3), consistent with a role for CDK2 in regulating the onset of mitosis as previously reported.33 A more detailed analysis by three-color flow cytometry assessed the impact of the full range of CVT-313 concentrations on S phase progression, pHH3, and DNA damage (Figure 3). Addition of CHK1i to SN38-arrested cells induced pHH3 in 14% of cells, in the G2/M phase; pHH3 was inhibited at low concentrations of CVT-313 (50% inhibition at 0.6 μM). WEE1i dramatically increased pHH3 in 70% of cells, but this was also inhibited by 50% at 1.25 μM CVT-313. In contrast, far higher concentrations of CVT-313 (40–80 μM) were required to prevent S phase progression (Figure 3).
Figure 3.
High concentrations of CDK1/2i prevent abrogation of the S phase, while low concentrations prevent mitotic entry. MDA-MB-231 cells were incubated with SN38 for 24 h and then CHK1i or WEE1i from 24 to 30 h. Additionally, CDK1/2i or CDC7i was administered concurrently with CHK1i or WEE1i as indicated. Cells were immunostained with fluorescent anti-pHH3 and anti-γH2AX antibodies, and DNA was stained with propidium iodide and then analyzed by flow cytometry. The gates and percentages shown represent the proportion of cells positive for pHH3 or γH2AX.
To assess the impact of CHK1i and WEE1i on DNA damage in SN38-arrested cells, we probed lysates for phosphorylation of the 32 kDa subunit of replication protein A (RPA32) and γH2AX. Both CHK1i and WEE1i increased γH2AX 6 h after administration compared to that with SN38 alone (Figure 2a). We also observed several bands with a reduced electrophoretic mobility when probing with the total RPA32 antibody, consistent with ATR- and DNA-PK-mediated phosphorylation.34 The slowest-mobility band corresponded to phosphorylation on serines 4 and 8, the DNA-PK site.34 Furthermore, the appearance of γH2AX coincided with induction of pHH3; WEE1i caused greater induction of both markers compared to CHK1i. Furthermore, CDC25i and CDK1/2i suppressed γH2AX and pRPA32 signals back to the level of SN38 alone (Figures 2a and 3).
Addition of CDC7i to WEE1i-treated cells prevented S phase progression (Figures 1b and 3) but surprisingly did not prevent increases in pHH3 and γH2AX (Figures 2a and 3). The increase in pHH3 occurred in the S phase-arrested cells but was again prevented by CVT-313 at low concentrations (Figure 3). These results suggest that the cells are undergoing premature mitosis from the S phase; thus, S phase progression is not necessary for WEE1i-mediated sensitization to SN38. This combination is studied further below. It is important to note that CHK1i and CDC7i did not increase pHH3 or γH2AX (Figure 2a), so while these cells remain arrested in the S phase (Figure 1c), they did not undergo premature mitosis, at least within this 6 h period. The regulation of mitotic entry by CDK1 or CDK2 is investigated further below.
Kinetics of Cell Cycle Progression and Mitosis Induced by SN38 with CHK1i or WEE1i
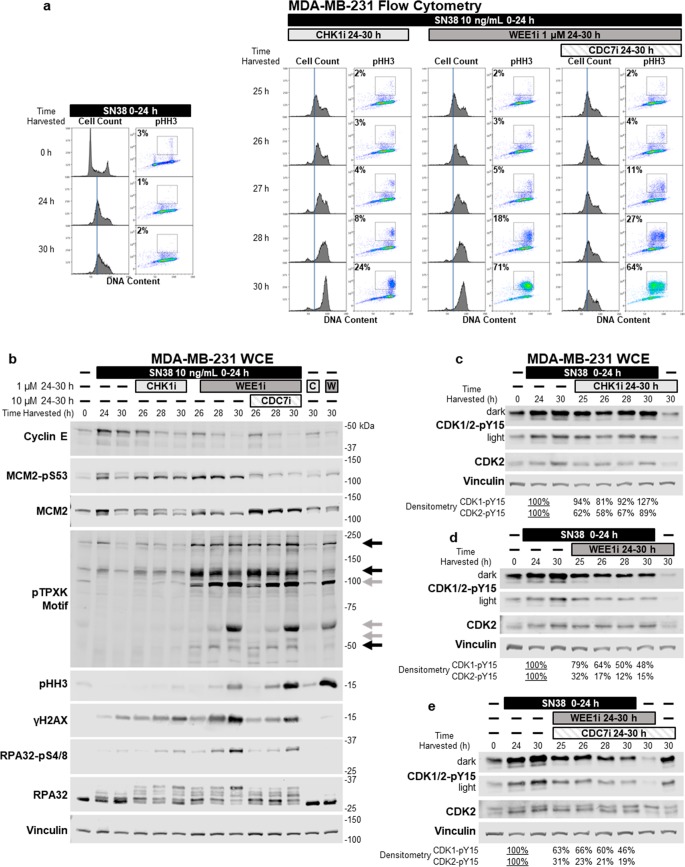
We assessed the kinetics of S phase progression and mitotic entry to investigate if either event coincided with an increased level of DNA damage following CHK1i and WEE1i. Following SN38-mediated arrest, MDA-MB-231 cells were incubated with CHK1i, WEE1i, or the combination of WEE1i and CDC7i, harvested after 1–6 h, and analyzed by flow cytometry (Figure 4a). S phase progression began within 2 h following CHK1i, but pHH3 increased only after 4–6 h (28–30 h post-SN38) as the cells reached the G2/M phase. WEE1i elicited similar kinetics but with many more cells positive for pHH3. With the combination of WEE1i and CDC7i, the onset of pHH3 was also delayed by 4–6 h but, as described above, occurred predominately in S phase cells.
Figure 4.
Kinetics of CHK1i- and WEE1i-mediated activation of CDK1/2, induction of mitotic entry, and DNA damage upon addition to SN38-arrested cells. (a) MDA-MB-231 cells were incubated with SN38 for 24 h, CHK1i or WEE1i added, and cells harvested at 25–30 h. Cells were immunostained with a fluorescent anti-pHH3 antibody, and DNA was stained with propidium iodide and then analyzed by flow cytometry. The gates and percentages shown represent the proportion of cells positive for pHH3. The vertical line defines the peak of cells at 24 h to emphasize progression visible upon addition of CHK1i or WEE1i. (b) MDA-MB-231 cells were incubated with SN38 for 24 h and then with CHK1i or WEE1i and harvested at 26–30 h. Additionally, CDC7i was administered concurrently with WEE1i as indicated. Whole cell extracts were probed for the indicated proteins by Western blotting. Black arrows indicate bands that are phosphorylated by 2 h following CHK1i or WEE1i, and gray arrows indicate bands phosphorylated around 6 h; these arrows highlight the same bands as in Figure 2a. (c–e) Incubations as in panels a and b but with cell lysates being assessed for phosphorylation of CDK1 and CDK2 using fluorescent secondary antibodies and a fluorescent scanner. Values reflect the average densitometry from three to six experiments; the individual values and statistical significance are presented in Figure S2.
Parallel Western blots demonstrated that cyclin E decreased rapidly following CHK1i or WEE1i (Figure 4b), indicating rapid activation of CDK2. However, γH2AX and pHH3 did not increase until 28–30 h (Figure 4b), consistent with flow cytometry results (Figure 4a). Increased phosphorylation of RPA32 occurred rapidly, but the appearance of the slowest-migrating band that is also phosphorylated on S4/S8 was also delayed to 28–30 h (Figure 4b). Intriguingly, almost all RPA32 is phosphorylated at 30 h in cells treated with SN38 and WEE1i. The lack of unphosphorylated RPA32 suggests that cells have extensive single-stranded DNA, probably exceeding cellular pools of RPA as we previously observed in cells incubated with gemcitabine and CHK1i;3 mitotic entry with excessive ssDNA likely contributes to the enhanced cytotoxicity of WEE1i in SN38-arrested cells. Similar results were observed in HeLa cells by flow cytometry and Western blots except that SN38 treatment alone resulted in significant phosphorylation of RPA32 (Figure S1). Addition of WEE1i depleted the residual unphosphorylated form of RPA32 and induced γH2AX and pHH3.
Resolution of the Activation of CDK2 and CDK1
We have previously discussed the difficulty of discriminating between phospho forms of CDK1 and CDK2 because the inhibitory phosphotyrosine 15 resides in the middle of a conserved 13-amino acid sequence.25 We recently demonstrated that CDK1-pY15 and CDK2-pY15 can be electrophoretically resolved on a Western blot when using fluorescent secondary antibodies and a fluorescent scanner.3 Furthermore, the increased dynamic range of fluorescent detection has made it possible to reliably quantify the 10-fold greater amount of CDK1-pY15 than of CDK2-pY15 (Figure 4c–e and Figure S2). Intriguingly, addition of CHK1i to S phase-arrested cells rapidly dephosphorylated CDK2-pY15 but had no impact on CDK1-pY15. Dephosphorylation of CDK2-pY15 was only partial (∼40%) (Figure 4c and Figure S2), which is consistent with prior reports that CDK2 is phosphorylated before binding to cyclin, and much of it remains unbound (Figure 2b).4,29 Presumably, CDC25A can dephosphorylate CDK2 only when complexed to a cyclin, thereby explaining the residual CDK2-pY15. In contrast, WEE1i caused almost complete dephosphorylation of CDK2 (Figure 4d,e and Figure S2), which can be attributed to WEE1i preventing phosphorylation of both cyclin-bound and -unbound CDK2. In addition, WEE1i caused significant but gradual dephosphorylation of CDK1 over 6 h (Figure 4d,e and Figure S2).
Analysis of CDK substrates was visualized with an antibody recognizing the CDK1/2 consensus sequence, pTPXK. There was rapid phosphorylation of some substrates at 26 h, such as the bands at 200, 120, and 50 kDa (Figure 4b, black arrows). However, additional substrates were selectively phosphorylated at 30 h, including the bands at 100, 65, and 55 kDa (gray arrows). The delayed appearance of these bands coincides with induction of pHH3 and a decrease in CDK1-pY15. Additionally, comparison of the time course (Figure 4b) and CVT-313 titration experiments (Figure 2a) shows that the early phosphorylated bands are inhibited at high concentrations of CVT-313, which prevent S phase progression. Furthermore, the delayed bands are inhibited with lower concentrations of CVT-313 that correlate with inhibition of mitosis. We conclude that the early phosphorylated bands are CDK2 substrates while the delayed bands are CDK1 substrates.
CDC25i, CDK1/2i, and CDC7i Prevent Markers of DNA Helicase Activity
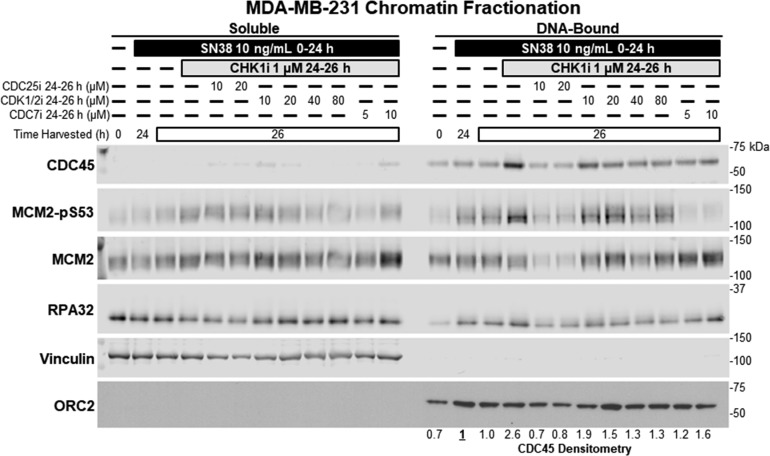
CDK2 functions in the S phase by phosphorylating treslin, which then mediates loading of CDC45 onto DNA as the rate-limiting step in origin firing.6 Chromatin fractionation was performed following drug treatments, with vinculin and ORC2 used as controls to confirm separation of these two compartments (Figure 5). Upon incubation with SN38, there was a small increase in the level of chromatin-bound CDC45 perhaps because more cells are in the S phase. Similarly, there is an increase in the level of DNA-bound MCM2-pS53, again consistent with CDC7 remaining active during arrest.
Figure 5.
CDC25i, high concentrations of CDK1/2i, and CDC7i prevent loading of CDC45 on chromatin. MDA-MB-231 cells were incubated with SN38 for 24 h and then with CHK1i for an additional 2 h. CDC25i, CDK1/2i, or CDC7i were administered concurrently with CHK1i as indicated. Soluble and chromatin-bound proteins were separated and probed for the indicated proteins by Western blotting.
Within 2 h of addition of CHK1i, there was a clear increase in the amount of CDC45 bound to DNA as well as an increase in the amount of faster-migrating phosphorylated form of MCM2-pS53 (Figure 5). This is consistent with induction of S phase progression within 2 h of CHK1i (Figure 4a). CDC25i resulted in a robust decrease in the levels of DNA-bound MCM2 and CDC45, suggesting a broad impact on replication firing. CDK1/2i inhibition also decreased CDC45 loading but had a weaker impact on MCM2 compared to CDC25i; the suppression of CDC45 loading was most pronounced at higher concentrations of CVT-313 (40–80 μM), which also inhibited S phase progression. Additionally, CDC7i caused dephosphorylation of MCM2 and prevented the CHK1i-mediated increase in the loading of CDC45 onto chromatin.
WEE1i with SN38 Is More Cytotoxic Than CHK1i with SN38
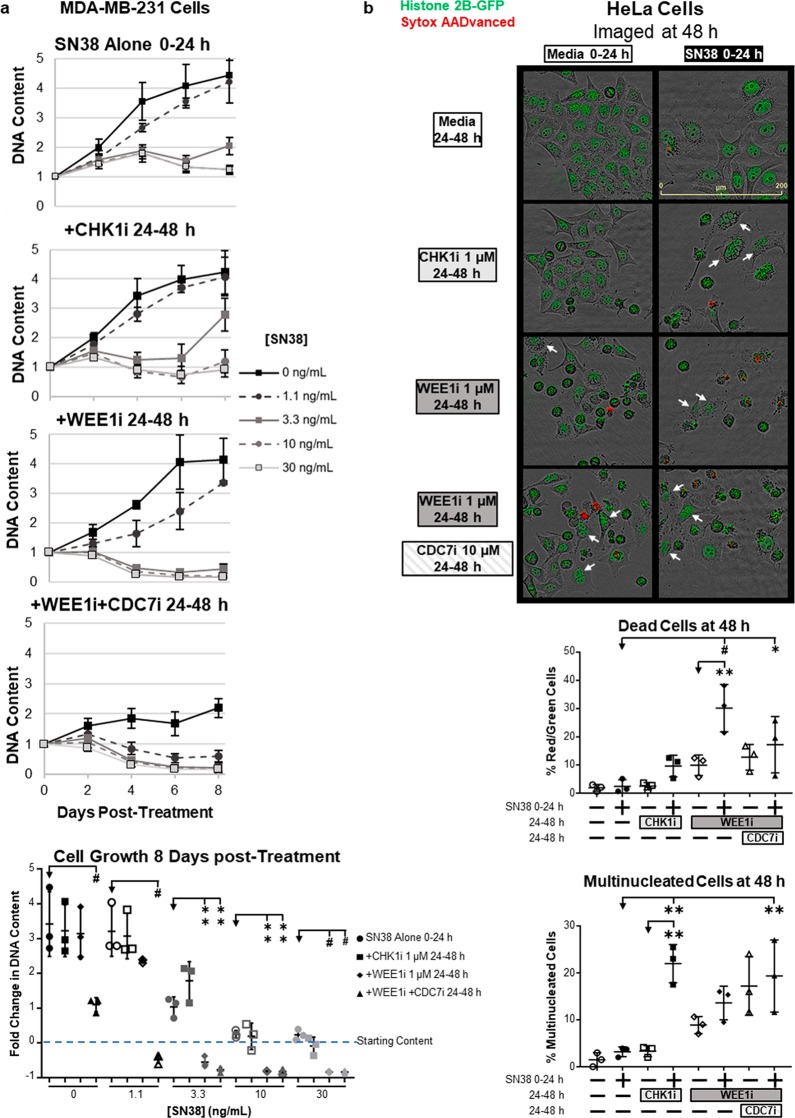
To investigate the effect of the drug combinations on cytotoxicity, we measured DNA content over 8 days following treatment (Figure 6a). With SN38 alone, 10–30 ng/mL resulted in cytostasis, i.e., no significant increase in DNA content by day 8. CHK1i resulted in a slight decrease in DNA content on days 4 and 6, but cells recovered to the SN38-alone value by day 8. In contrast, WEE1i elicited robust cytotoxicity when combined with >3.3 ng/mL SN38 with near complete elimination of cells by day 8. Additionally, WEE1i and CDC7i without SN38 significantly slowed cell growth and dramatically sensitized cells to even 1.1 ng/mL SN38. These data demonstrate that WEE1i more effectively sensitizes cells to SN38 than CHK1i, likely due to it directly preventing the inhibitory phosphorylation on CDK1-Y15.
Figure 6.
WEE1i, but not CHK1i, significantly sensitize cells to SN38. (a) MDA-MB-231 cells were incubated with SN38 for 24 h and then with 1 μM CHK1i or WEE1i from 24 to 48 h. Additionally, 10 μM CDC7i was administered concurrently with WEE1i as indicated. Separate plates were harvested every 2 days. DNA content was assessed and normalized to the starting value (day 0). Line graphs indicate means ± the standard deviation (SD) of DNA content (n = 3). The scatter plot represents the mean ± SD for the change in DNA content at day 8. **p < 0.005, and #p < 0.0001 (n = 3). (b) HeLa cells stably expressing histone 2B-GFP were incubated as described for panel a. Cells were imaged by the Incucyte Zoom system at 48 h (see Figures S3 and S4 for the full time course). The cell membrane permeability and cell number were quantified by fluorescent signals. White arrows denote examples of multinucleated cells, which were counted manually. Scatter plots indicate the mean ± SD percentage of dead or multinucleated cells at 48 h. *p < 0.05, **p < 0.005, and #p < 0.0001 (n = 3).
We further characterized cell death by time lapse imaging with HeLa cells stably expressing histone 2B-GFP and in the presence of a membrane impermeant DNA stain. Cells were incubated with SN38 for 24 h, and then images were acquired every 2 h in the presence of CHK1i or WEE1i (between 24 and 48 h). Cells incubated with 10 ng/mL SN38 alone showed no increase in the number of nuclei, and only a few cells exhibited membrane permeability by 48 h (Figure 6b and Figures S3 and S4). Addition of CHK1i at 24 h caused most of the cells to round up by 32 h, consistent with them entering mitosis, but most spread out again and exhibited multinucleated cells by 48 h as cytokinesis failed; again, few cells exhibited membrane permeability. Addition of WEE1i or WEE1i with CDC7i also caused cells to round up, but subsequently, membrane integrity was lost by many more. These data suggest that potent activation of CDK1 by WEE1i significantly enhances cytotoxicity in SN38-arrested cells.
Impact of Different Concentrations of CDK1/2i in AsPC-1 Cells
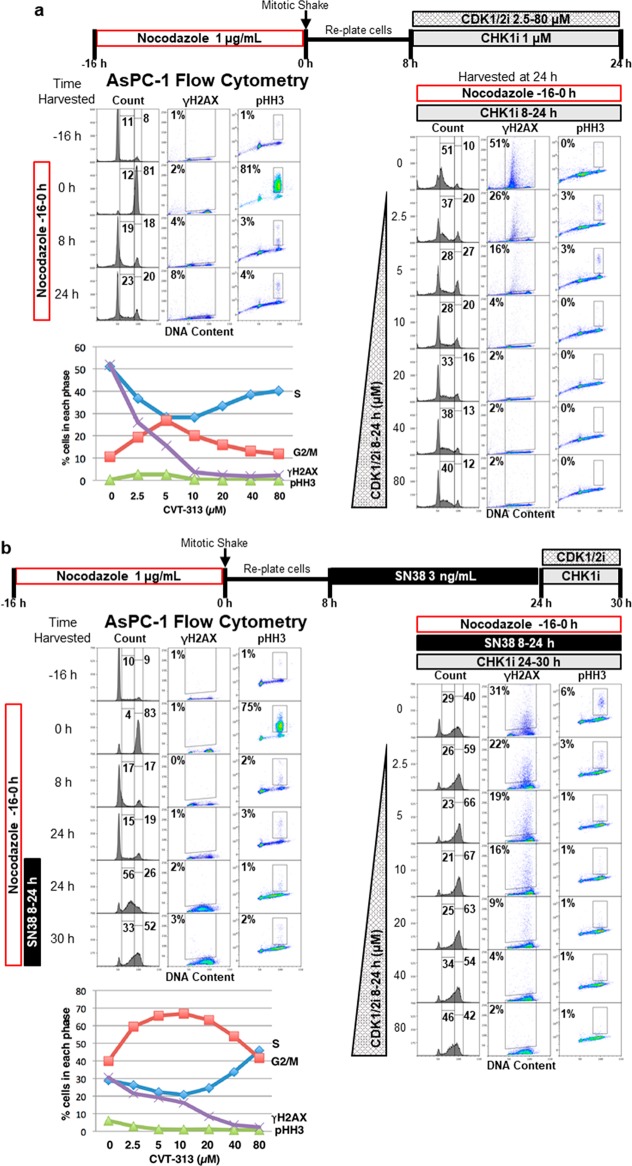
We next assessed the role of CDK2 in AsPC-1 cells to ensure the effects of CVT-313 were not unique to just one cell line. Furthermore, AsPC-1 cells are very sensitive to CHK1i as a single agent,4 which provided the opportunity to demonstrate the variable impact of CVT-313 concentration on cell cycle perturbation in a single model. AsPC-1 cells readily synchronize and recover from a 16 h incubation in nocodazole. The majority of cells re-entered G1 by 4 h after replating; by 12 h, the majority of cells were in early S phase (Figures 7 and 8 and Figure S5). Hence, this synchronized model can be used to discriminate progression through the G1, S, or G2 phase.
Figure 7.
Impact of different concentrations of CDK1/2i on CHK1i-induced γH2AX, S phase progression, and mitotic entry. (a) AsPC-1 cells were synchronized by incubation with nocodazole for 16 h. Mitotic cells were collected and replated in fresh medium for 8 h. CHK1i and CDK1/2i were administered from 8 to 24 h at the indicated concentrations. Cells were immunostained with fluorescent anti-pHH3 and anti-γH2AX antibodies, and DNA was stained with propidium iodide and then analyzed by flow cytometry. The gates, percentages, and line graph represent the proportion of cells positive for S phase DNA content, G2 DNA content, pHH3, or γH2AX. (b) Synchronized AsPC-1 cells were incubated with 3 ng/mL SN38 from 8 to 24 h after being released from nocodazole. CHK1i and CDK1/2i were administered at 24–30 h at the indicated concentrations. Cells were harvested and analyzed as described for panel a. A repeat of this experiment in cells incubated with a 5 ng/mL concentration is presented in Figure S5.
Figure 8.
Impact of the concentration of CDK1/2i and CDC7i in undamaged cells. AsPC-1 cells were synchronized by incubation with nocodazole for 16 h. Mitotic cells were collected and replated in fresh medium for 4 h (left) or 12 h (right). CDK1/2i or CDC7i were administered from 4 to 16 h (left) or from 12 to 24 h (right) at the indicated concentrations. DNA was stained with propidium iodide and then analyzed by flow cytometry.
Sensitivity to CHK1i as a single agent is observed as increased γH2AX in S phase cells (Figure 7).4 Accordingly, we added CHK1i to synchronized cells just prior to them entering the S phase (8 h after being released from nocodazole). At 24 h (incubation for 16 h with CHK1i), the majority of cells were arrested in the early S phase with extensive γH2AX (Figure 7). Concurrent incubation with 2.5–5 μM CVT-313 reduced γH2AX and overcame S phase arrest, resulting in an increase in the percentage of cells with G2/M DNA content and pHH3. At 10 μM CVT-313, there was a reduction in the percent of cells positive for pHH3, suggesting inhibition of CDK1. At higher concentrations (40–80 μM), cells again accumulated in the S phase, consistent with these higher concentrations preventing CDC45 loading and S phase progression.
We also assessed the impact of CVT-313 on cytotoxicity induced by CHK1i monotherapy (Figure S6). Low concentrations of CVT-313 (2.5 and 5 μM) did not inhibit proliferation but dramatically rescued cells from MK-8776-induced cytotoxicity. Higher concentrations of CVT-313 (10 μM) inhibited cell growth and therefore could not rescue cells from cytotoxicity. This was observed in both AsPC-1 and U2OS cells, both of which are sensitive to single agent MK-8776.4
We also titrated CVT-313 in the synchronized model of AsPC-1 cells incubated with SN38 and CHK1i. A low concentration of SN38 (3 ng/mL) added 8–24 h after release from nocodazole arrested most of the cells in the mid-S phase (Figure 7b). CHK1i partially abrogated S phase arrest, although incompletely as the single-agent sensitivity of AsPC-1 induced additional γH2AX. Concurrent incubation with low concentrations of CVT-313 reduced γH2AX and enhanced abrogation of S phase arrest, and more cells accumulated in the G2/M phase. At the higher concentrations of CVT-313, inhibition of S phase progression was again observed. This experiment was repeated with 5 ng/mL SN38 with similar conclusions but less abrogation of S phase arrest by CHK1i with CDK2i (Figure S5). There is only a small concentration range of SN38 over which all of these events can be observed as CHK1i failed to abrogate arrest induced by higher concentrations of SN38 due to the cumulative impact of both the single agent and combination effects.
Both of these experimental strategies demonstrated that low concentrations of CVT-313 suppress γH2AX induced by CHK1i either as a single agent or in combination with SN38 and facilitate S phase progression. Suppression of γH2AX is consistent with our prior report that CVT-313 prevented double-strand breaks induced by CHK1i monotherapy.4 In contrast, higher concentrations of CVT-313 prevent S phase progression and mitosis. While the induction of mitosis is generally considered a result of CDK1 activity, both the monotherapy activity and S phase progression appear to depend on CDK2 but at very different concentrations of CVT-313.
Impact of CDK1/2i and CDC7i in Undamaged Cells
Finally, we questioned whether high concentrations of CVT-313 were also required to prevent S phase progression in undamaged cells. We used the synchronized AsPC-1 model to discriminate progression through the G1, S, or G2 phase. CVT-313 was added from 4 to 16 h to assess the impact on the G1 to S phase transition or from 12 to 24 h to assess the progression of cells from the S to G2 phase. Addition of 5–10 μM CVT-313 arrested cells in the G1 phase (Figure 8). In contrast, 5 μM CVT-313 failed to prevent progression of cells from the S to G2 phase. However, 5 μM CVT-313 reduced the number of cells undergoing mitosis, perhaps through inhibition of CDK1. The highest concentrations of CVT-313 (40–80 μM) only partially retarded S phase progression. As the cells have no exogenous damage, this suggests that ongoing replicons can continue and that CDK2 may be primarily required to fire dormant origins when some of the replicons stall from endogenous damage.
In parallel, we assessed the impact of CDC7i on entry into and progression through the S phase in undamaged cells. Surprisingly, CDC7i arrested cells in the early S phase rather than the G1 phase, suggesting that entry into the S phase might be able to circumvent the requirement for CDC7 (Figure 8). CDC7i was also a poor inhibitor of S phase progression when added from 12 to 24 h after release from nocodazole, but like CVT-313, S phase progression was slowed or incomplete, suggesting CDC7 was required for completion of the S phase. Overall, these results suggest that CDC7 and CDK2 are both essential for S phase progression, but CDK2 is primarily responsible for S phase initiation.
Discussion
Having previously established that CDK2 activity in the S phase is required for cytotoxicity induced by CHK1i as a single agent,4 we first sought in this study to understand how CHK1i can also abrogate S phase arrest in SN38-treated cells, which is also well-established to require CDK2 activity. With this goal in mind, we demonstrated that different levels of CDK2 activity determine the molecular mechanism elicited by CHK1i. These experiments relied on CVT-313, which is 10-fold selective for CDK2 over CDK1 against purified kinases.21 Our results demonstrated that the different CDK2-dependent effects are prevented with different concentrations of CVT-313. In AsPC-1 cells, CHK1i-mediated induction of γH2AX and cytotoxicity were suppressed by low concentrations of CVT-313, which consequently enhanced S phase progression (Figure 7 and Figure S6). Intermediate concentrations of CVT-313 prevented progression from the G1 to S phase in undamaged cells, while much higher concentrations were required to prevent S phase progression in damaged cells as well as completion of the S phase in undamaged cells.
As CVT-313 also inhibits CDK1 and mitosis at intermediate concentrations (10 μM), our observations raised the question of whether all of the other events, and particularly the prevention of S phase progression at high concentrations of CVT-313, can still be attributed to inhibition of CDK2. The following results support this conclusion: selective dephosphorylation of CDK2 but not CDK1 upon addition of CHK1i (Figure 6b and Figure S2) and high concentrations of CVT-313 were required to prevent degradation of cyclin E, as well as chromatin loading of CDC45, both effects normally attributed to CDK2 activity.6,26,27 This study (Figure 2a) and our recent publication on the combination of gemcitabine and CHK1i3 further demonstrate that phosphorylation of different CDK substrates is differentially sensitive to CVT-313. Our prior study also confirmed that high concentrations of CVT-313 prevented CHK1i-mediated CDC45 chromatin loading. Additionally, the ability of a pan CDC25i to prevent CDC45 loading and S phase progression further supports the conclusion that CHK1 signaling through the canonical pathway of CDC25 and CDK2 is responsible for causing S phase arrest following SN38 treatment. These observations suggest that phosphorylation of critical substrates requires different levels of CDK2 activity, and this underlies the molecular mechanisms that discriminate CHK1i activity as a single agent from its activity in drug combinations.
An explanation for these observations can be found in experiments using a monomolecular CDK/cyclin module in Schizosaccharomyces pombe.18,19 Those experiments demonstrated that CDK activity increases as cells pass through the S and G2 phases and that CDK activity thresholds exist for specific substrates, thereby providing temporal ordering of the S phase and mitosis. The sensitivity of the various substrates to CDK activity was also reflected in the concentration of CDKi required to inhibit their phosphorylation, with S phase substrates being ≤1000-fold more resistant to CDKi than M phase substrates.19 This is consistent with our observations that S phase effects (e.g., CDC45 loading and S phase progression) are the most resistant to CVT-313 and therefore support the hypothesis that these are indeed CDK2-dependent.
A clear problem with this study is the lack of truly selective inhibitors for CDK2. In addition to CDK1/2, CVT-313 also inhibits CDK5 at similar concentrations. CDK5 has many reported roles in neuronal maturation, insulin secretion, and PD-L1 expression, but evidence of its involvement in cell cycle regulation is very tangential.35 Therefore, it is unlikely that inhibition of CDK5 is responsible for CVT-313-mediated S phase arrest at high concentrations. We are unaware of any other inhibitor that exhibits selectivity for CDK2 over alternate CDKs in cells; for example, while NU6140 was reported to be selective for CDK2, it arrested cells in the G2/M phase rather than the G1 phase, which is inconsistent with the role of CDK2.36 Genetic approaches such as RNAi or CRISPR.Cas9 to suppress CDK2 induce G1 arrest and consequently confer resistance to DNA-damaging agents that affect S phase cells.37−39 Contradictory literature has demonstrated that cells lacking CDK2 are viable and continue to proliferate and mice lacking CDK2 are also viable,40,41 but this is explained by the redundancy of CDK1 and CDK2. However, when both kinases are present, they clearly have many different functions. The advantage of pharmacologic inhibitors is that they can be added concurrently with CHK1i and immediately inhibit the desired target in cells.
We also used small molecule inhibitors to show the importance of CDC7 for abrogation of S phase arrest, but the specificity of such inhibitors has also raised concerns. XL413 was discovered as a potent and selective inhibitor of CDC7;42 however, it was subsequently reported that XL413 had limited activity in most cells, and this was hypothesized to be due to limited bioavailability.43 The fact that XL413 strongly inhibited MCM2 phosphorylation suggests that, at least in the cell models used here, it is effectively inhibiting CDC7. It has been suggested that PHA-767491 is a better CDC7 inhibitor, but this compound is also an inhibitor of CDK9, which results in global inhibition of transcription.44 Hence, results obtained with PHA-767491 must be seriously questioned.
In this study, we show that SN38-induced phosphorylation of MCM2-S53 was prevented by XL413 (Figures 2a, 4b, and 5 and Figure S1). XL413 also prevented S phase progression (Figures 1c, 3, 4, and 8 and Figure S1). These data demonstrate that CDC7 is required for reactivation of the DNA replication machinery following DNA damage-mediated arrest. In contrast, XL413 poorly inhibited S phase progression in undamaged cells (Figure 8). Furthermore, in undamaged cells, XL413 failed to prevent progression from the G1 to S phase; rather, the cells were arrested in the early S phase (Figure 8). The original discovery of XL413 also demonstrated arrest in the late S phase rather than the G1 phase as expected from its proposed role in replication origin firing.42 The failure of XL413 to inhibit early origin firing contradicts findings from siRNA knockdown of CDC7 that did prevent the G1 to S phase transition in IMR90 fibroblasts.45 The role of CDC7 in regulating firing of early origins remains unclear, but its role in dormant origin firing after S phase arrest is supported by our observations.
This study also compared the efficacy of CHK1i and WEE1i in promoting SN38-mediated cytotoxicity. We previously demonstrated that CHK1i promoted aberrant mitotic entry after cells had progressed to the G2 phase following SN38 arrest.14,20,46 The study presented here demonstrates that WEE1i more effectively sensitized cells to SN38. The enhanced cytotoxicity mediated by WEE1i correlated with a robust decrease in CDK1-pY15 and a greater induction of pHH3. This is likely due to the direct regulation of CDK1 by WEE1, in contrast to CHK1 indirectly regulating CDK1 through the upstream CDC25 phosphatases. Cells appeared to enter mitosis around 6–8 h following CHK1i or WEE1i. Interestingly, the cells subsequently re-adhered by 24 h following CHK1i with fragmented nuclei. The re-adherence, despite nuclear fragmentation, appeared to be at least a transient survival mechanism as the culture remained cytostatic for 8 days. Conversely, more cells lost membrane integrity 24 h following WEE1i, and there was a marked loss of viable cells by 8 days. The combination of WEE1i and CDC7i following SN38 also induced extensive cytotoxicity over 8 days, but the mitotic events occurred while the cells were still in the S phase.
A potentially confounding factor of using AZD1775 to inhibit WEE1 is that PLK1 is also inhibited at similar concentrations, at least in in vitro kinase assays.47 However, PLK1 activity is well characterized to promote mitotic entry through negatively regulating WEE1 and activating CDC25C.48,49 Consequently, inhibition of WEE1 would counteract any inhibition of PLK1, and indeed, our data demonstrate that AZD1775 potently induces mitotic entry of SN38-damaged cells. Therefore, inhibition of PLK1 does not appear to prevent mitotic entry. However, preventing the ability of PLK1 to stabilize the mitotic spindle may exacerbate mitotic defects and promote apoptosis.50 Further investigation could characterize the contribution of PLK1 to the induction of mitotic catastrophe in the context of topoisomerase inhibitor treatment.
A likely contributing factor to cytotoxicity was the extensive ssDNA following CHK1i or WEE1i, as demonstrated by phosphorylation of RPA32. In both MDA-MB-231 and HeLa cell lines, there remained little if any unphosphorylated RPA at the time they entered mitosis after incubation with WEE1i. This suggests that the extent of ssDNA exceeds the protective capacity of RPA. Excessive ssDNA likely interferes with chromosomal segregation and confers instability to chromosomal structure. We conclude that WEE1 inhibitors would be better candidates for clinical trials investigating combination therapies with topoisomerase inhibitors, but whether they would also be more toxic to the patient remains to be determined. A notable exception is the reported synthetic lethality of tumors with RAD50 mutations upon their treatment with a topoisomerase inhibitor with CHK1i.51 There may be other specific situations in which the combination with a CHK1i would be very selective for a tumor.
Materials and Methods
Cell Culture
MDA-MB-231 cells were obtained from the Developmental Therapeutics Program of the National Cancer Institute. AsPC-1 and U2OS cells were obtained from ATCC. HeLa cells stably expressing histone 2B-GFP were a generous gift from A. Kettenbach (Geisel School of Medicine). Cells were cultured as described previously.3
Small Molecule Inhibitors
SN38 (Pfizer, New York, NY), CVT-313 (Sigma-Aldrich), MK-8776 (Merck, Kenilworth, NJ), AZD1775 (Merck), and NSC663284 (Cayman Chemical, Ann Arbor, MI) were dissolved in dimethyl sulfoxide. XL413 (Tocris, Bristol, U.K.) was dissolved in water.
Antibodies
The following antibodies were used at the indicated dilutions.
Abcam (Cambridge, MA): RPA32 (1:5000; RRID, AB_302873).
Bio-Rad (Hercules, CA): anti-mouse HRP (1:3000; RRID, AB_11125547) and anti-rabbit HRP (1:3000; RRID, AB_11125547).
Cell Signaling (Danvers, MA): anti-mouse DyLight680 (1:20000; RRID, AB_10696895), anti-mouse DyLight800 (1:20000; RRID, AB_10693543), anti-rabbit DyLight800 (1:20000; RRID, AB_10697505), CDK1/2-pY15 (1:1000; RRID, AB_331460), CDK substrates/pTPXK (1:1000, catalog no. 14371), H2AX-pS139 (1:2000; RRID, AB_2118009), H2AX-pS139-Alexa488 (1:100; RRID, AB_10694488), HH3-pS10 (1:3000; RRID, AB_1549592), HH3-pS10-Alexa647 (1:200; RRID, AB_1549592), MCM2 (1:5000; RRID, AB_2687884), ORC2 (1:1000; RRID, AB_10694717), and PCNA-Alexa488 (1:1000; RRID, AB_11178664).
Novus Biologicals (Littleton, CO): RPA-pS4/8 (1:3000; RRID, AB_1726226).
Millipore (Burlington, MA): CDK2 (1:1000; RRID, AB_2291613).
Santa Cruz Biotechnology (Santa Cruz, CA): CDC45 (1:3000; RRID, AB_2078507), cyclin E (1:1000; RRID, AB_627357), and vinculin (1:3000; RRID, AB_1131294).
Bethyl Laboratories (Montgomery, TX): MCM2-pS53 (1:3000; RRID, AB_669843).
Sigma-Aldrich: actin-HRP (1:100000; RRID, AB_262011).
Thermo Fisher Scientific (Carlsbad, CA): anti-rat HRP (1:1000; RRID, AB_2535648).
Western Blotting
Cells were plated at densities of 2–300000 per well in a six-well plate. Following treatment, cells were harvested, and the proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed with primary and secondary antibodies as previously described.3 Blots in Figure 2b and ORC2 in Figure 5 were developed by chemiluminecense and X-ray film. All other blots were imaged with fluorescent antibodies on the Licor Odessey Clx instrument (Licor Biosciences, Lincoln, NE), which provides a significantly wider dynamic range. Densitometry of CDK1/2-pY15 blots was performed with GelBandFitter.52
Chromatin Fractionation
Two million cells per 10 cm dish were plated, treated, suspended with a cell scraper, and transferred to a 15 mL tube. Soluble and chromatin-bound proteins were separated as previously described3 and then analyzed by Western blotting as described above. Vinculin was used as a soluble fraction control, and ORC2 was used as a DNA-bound control.
Immunoprecipitation
Cells were lysed in lysis/wash buffer provided in the Classic Magnetic IP/Co-IP Kit (Pierce, 8804) with added protease and phosphatase inhibitors for 30 min on ice and then centrifuged at 13000g for 10 min. The extract (500 μg) was precleared with Protein A/G magnetic beads for 1 h at 4 °C, and the supernatant was collected. The antibody [2.5 μg; cyclin E (sc-198)] was added to the supernatant and mixed for 3 h at 4 °C. Prewashed magnetic beads were added and incubated on a rotator at 4 °C overnight. The supernatant was recovered, and the immunoprecipitate was washed and resuspended in 2× Laemmli sample buffer. Equivalent portions of the supernatant and immunoprecipitate were analyzed by Western blotting as described above.
Flow Cytometry
Cells were plated at densities of 2–300000 per well in a six-well plate. Following treatment, cells were harvested and stained for γH2AX, pHH3, and DNA as previously described.3 Cells were analyzed on the Gallios flow cytometer (Beckman Coulter, Brea, CA), and the data were analyzed using FlowLogic software.
Time Lapse Microscopy
HeLa cells expressing histone 2B-GFP were plated at a density of 2000 cells per well in a 96-well plate and treated with 10 ng/mL SN38 the following day (three wells/technical replicates per treatment condition). Twenty-four hours following SN38, cells were washed with phosphate-buffered saline, and then fresh medium, 1 μM MK-8776, 1 μM AZD1775, or 1 μM AZD1775 with 10 μM XL413 was added to the wells. Additionally, either 250 nM Cytotox Red (Essen Bioscience, Ann Arbor, MI) or 1 μM Sytox AAdvanced (Thermo Fischer) was used to monitor cell membrane permeability. Plates were imaged by the Incucyte Zoom (Essen Bioscience) system every 2 h until 48 h at 37 °C. The Incucyte Zoom software was used to calculate confluency and the number of green and red objects. Multinucleated cells were manually counted.
Cell Death Assay
Cells were plated at a density of 10000 cells per well in 96-well plates (eight wells per treatment condition). Separate plates were harvested on day 0 (before treatment) and on days 2, 4, 6, and 8 following treatment. DNA was stained with Hoechst 33258 and quantified as described previously.53,54
Quantification and Statistical Analysis
Quantitative data are shown as means ± the standard deviation. Values of n reported in the figure legends or identified by scatter plots indicate independent biological repeats. Statistical analysis was performed using one-way analyses of variance with Sidak’s multiple-comparison test in Graphpad Prism 7; significant differences were determined with a p ≤ 0.05 threshold.
Acknowledgments
This research was supported by National Institutes of Health Grant 2RO1CA117874 to A.E. and National Cancer Institute Cancer Center Support Grant 5P30 CA023108-37 to the Norris Cotton Cancer Center.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsptsci.9b00001.
Kinetics of cell cycle arrest and abrogation in HeLa cells (Figure S1), quantitation of densitometry for pCDK1 and pCDK2 (Figure S2), images from movies of HeLa cells undergoing aberrant mitosis and quantification of various events (Figures S3 and S4), impact of different concentrations of CVT-313 on DNA damage and S phase progression in AsPC-1 cells (Figure S5), and the ability of CVT-313 to rescue cells from sensitivity to CHK1i (Figure S6) (PDF)
Author Contributions
N.J.H.W. and A.E. contributed to study design and wrote the manuscript. K.L.D. contributed data on the resolution of phosphorylated CDK1/2. A.E. provided overall project supervision.
The authors declare no competing financial interest.
This article is made available for a limited time sponsored by ACS under the ACS Free to Read License, which permits copying and redistribution of the article for non-commercial scholarly purposes.
Supplementary Material
References
- Thompson R.; Eastman A. (2013) The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design. Br. J. Clin. Pharmacol. 76, 358–369. 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L. I.; Altmeyer M.; Rask M.-B.; Lukas C.; Larsen D. H.; Povlsen L. K.; Bekker-Jensen S.; Mailand N.; Bartek J.; Lukas J. (2013) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103. 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Warren N. J. H.; Eastman A. (2019) Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. J. Biol. Chem. 294, 1763–1778. 10.1074/jbc.RA118.005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurikar N.; Thompson R.; Montano R.; Eastman A. (2016) A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget 7, 1380–1394. 10.18632/oncotarget.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M.; Barbacid M. (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166. 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Jones R. M.; Petermann E. (2012) Replication fork dynamics and the DNA damage response. Biochem. J. 443, 13–26. 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- Montagnoli A.; Valsasina B.; Brotherton D.; Troiani S.; Rainoldi S.; Tenca P.; Molinari A.; Santocanale C. (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281, 10281–10290. 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Chuang L.-C.; Teixeira L. K.; Wohlschlegel J. A.; Henze M.; Yates J. R.; Méndez J.; Reed S. I. (2009) Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol. Cell 35, 206–216. 10.1016/j.molcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A.; Shevchenko A.; Shevchenko A.; Dunphy W. G. (2011) Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 193, 995–1007. 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam C. G.; Goins D.; Siefert J. C.; Clowdus E. A.; Sansam C. L. (2015) Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes Dev. 29, 555–566. 10.1101/gad.246827.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S. L.; Cappell S. D.; Tsai F.-C.; Overton K. W.; Wang C. L.; Meyer T. (2013) The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383. 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villerbu N.; Gaben A.-M.; Redeuilh G.; Mester J. (2002) Cellular effects of purvalanol A: A specific inhibitor of cyclin-dependent kinase activities. Int. J. Cancer 97, 761–769. 10.1002/ijc.10125. [DOI] [PubMed] [Google Scholar]
- Thompson R.; Montano R.; Eastman A. (2012) The Mre11 nuclease is critical for the sensitivity of cells to Chk1 inhibition. PLoS One 7, e44021. 10.1371/journal.pone.0044021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn E. A.; Ruth N. D.; Brown M. K.; Livingstone M.; Eastman A. (2002) Abrogation of the S phase DNA damage checkpoint results in S phase progression or premature mitosis depending on the concentration of 7-hydroxystaurosporine and the kinetics of Cdc25C activation. J. Biol. Chem. 277, 26553–26564. 10.1074/jbc.M202040200. [DOI] [PubMed] [Google Scholar]
- Syljuasen R. G.; Sorensen C. S.; Hansen L. T.; Fugger K.; Lundin C.; Johansson F.; Helleday T.; Sehested M.; Lukas J.; Bartek J. (2005) Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25, 3553–3562. 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N.; Dutta A. (2011) The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 25, 621–633. 10.1101/gad.2029711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H.; Nähse V.; Larsen M. S. Y.; Groth P.; Clancy T.; Lees M.; Jørgensen M.; Helleday T.; Syljuåsen R. G.; Sørensen C. S. (2010) Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J. Cell Biol. 188, 629–638. 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse D.; Nurse P. (2010) Driving the cell cycle with a minimal CDK control network. Nature 468, 1074–1079. 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- Swaffer M. P.; Jones A. W.; Flynn H. R.; Snijders A. P.; Nurse P. (2016) CDK substrate phosphorylation and ordering the cell cycle. Cell 167, 1750–61.e16. 10.1016/j.cell.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano R.; Chung I.; Garner K. M.; Parry D.; Eastman A. (2012) Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol. Cancer Ther. 11, 427–438. 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks E. E.; Gray N. S.; Joly A.; Kerwar S. S.; Lum R.; Mackman R. L.; Norman T. C.; Rosete J.; Rowe M.; Schow S. R.; Schultz P. G.; Wang X.; Wick M. M.; Shiffman D. (1997) CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J. Biol. Chem. 272, 29207–29211. 10.1074/jbc.272.46.29207. [DOI] [PubMed] [Google Scholar]
- Aarts M.; Sharpe R.; Garcia-Murillas I.; Gevensleben H.; Hurd M. S.; Shumway S. D.; Toniatti C.; Ashworth A.; Turner N. C. (2012) Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discovery 2, 524–539. 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- Duda H.; Arter M.; Gloggnitzer J.; Teloni F.; Wild P.; Blanco M. G.; Altmeyer M.; Matos J. (2016) A mechanism for controlled breakage of under-replicated chromosomes during mitosis. Dev. Cell 39, 740–755. 10.1016/j.devcel.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Lazo J. S.; Aslan D. C.; Southwick E. C.; Cooley K. A.; Ducruet A. P.; Joo B.; Vogt A.; Wipf P. (2001) Discovery and biological evaluation of a new family of potent inhibitors of the dual specificity protein phosphatase Cdc25. J. Med. Chem. 44, 4042–4049. 10.1021/jm0102046. [DOI] [PubMed] [Google Scholar]
- Sakurikar N.; Eastman A. (2016) Critical reanalysis of the methods that discriminate the activity of CDK2 from CDK1. Cell Cycle 15, 1184–1188. 10.1080/15384101.2016.1160983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M.; Singer J.; Loeb K. R.; Grim J.; Bloecher A.; Gurien-West M.; Clurman B. E.; Roberts J. M. (2003) Multisite phosphorylation by Cdk2 and GSK3 controls Cyclin E degradation. Mol. Cell 12, 381–392. 10.1016/S1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- Clurman B. E.; Sheaff R. J.; Thress K.; Groudine M.; Roberts J. M. (1996) Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 10, 1979–1990. 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- Aleem E.; Kiyokawa H.; Kaldis P. (2005) Cdc2–cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7, 831–836. 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- Coulonval K.; Bockstaele L.; Paternot S.; Roger P. P. (2003) Phosphorylations of cyclin-dependent kinase 2 revisited using two-dimensional gel electrophoresis. J. Biol. Chem. 278, 52052–52060. 10.1074/jbc.M307012200. [DOI] [PubMed] [Google Scholar]
- Lee A. Y.-L.; Chiba T.; Truong L. N.; Cheng A. N.; Do J.; Cho M. J.; Chen L.; Wu X. (2012) Dbf4 is direct downstream target of Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J. Biol. Chem. 287, 2531–2543. 10.1074/jbc.M111.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenca P.; Brotherton D.; Montagnoli A.; Rainoldi S.; Albanese C.; Santocanale C. (2007) Cdc7 Is an active kinase in human cancer cells undergoing replication stress. J. Biol. Chem. 282, 208–215. 10.1074/jbc.M604457200. [DOI] [PubMed] [Google Scholar]
- Yamada M.; Watanabe K.; Mistrik M.; Vesela E.; Protivankova I.; Mailand N.; Lee M.; Masai H.; Lukas J.; Bartek J. (2013) ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 27, 2459–2472. 10.1101/gad.224568.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J.; Enders G. H. (2004) Cyclin A/Cdk2 complexes regulate activation of Cdk1 and Cdc25 phosphatases in human cells. Oncogene 23, 3361–3367. 10.1038/sj.onc.1207446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Opiyo S. O.; Manthey K.; Glanzer J. G.; Ashley A. K.; Amerin C.; Troksa K.; Shrivastav M.; Nickoloff J. A.; Oakley G. G. (2012) Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 40, 10780–10794. 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupp A.; Casimiro M. C.; Pestell R. G. (2017) Biological functions of CDK5 and potential CDK5 targeted clinical treatments. Oncotarget 8, 17373–17382. 10.18632/oncotarget.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennati M. (2005) Potentiation of paclitaxel-induced apoptosis by the novel cyclin-dependent kinase inhibitor NU6140: a possible role for survivin down-regulation. Mol. Cancer Ther. 4, 1328–1337. 10.1158/1535-7163.MCT-05-0022. [DOI] [PubMed] [Google Scholar]
- Du J.; Widlund H. R.; Horstmann M. A.; Ramaswamy S.; Ross K.; Huber W. E.; Nishimura E. K.; Golub T. R.; Fisher D. E. (2004) Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6, 565–576. 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Neganova I.; Vilella F.; Atkinson S. P.; Lloret M.; Passos J. F.; von Zglinicki T.; O’Connor J.-E.; Burks D.; Jones R.; Armstrong L.; Lako M. (2011) An important role for CDK2 in G1 to S Checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells 29, 651–659. 10.1002/stem.620. [DOI] [PubMed] [Google Scholar]
- Molenaar J. J.; Ebus M. E.; Geerts D.; Koster J.; Lamers F.; Valentijn L. J.; Westerhout E. M.; Versteeg R.; Caron H. N. (2009) Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. U. S. A. 106, 12968–12973. 10.1073/pnas.0901418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S.; Prieto I.; Odajima J.; Martín A.; Dubus P.; Sotillo R.; Barbero J. L.; Malumbres M.; Barbacid M. (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35, 25–31. 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Berthet C.; Aleem E.; Coppola V.; Tessarollo L.; Kaldis P. (2003) Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785. 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Koltun E. S.; Tsuhako A. L.; Brown D. S.; Aay N.; Arcalas A.; Chan V.; Du H.; Engst S.; Ferguson K.; Franzini M.; Galan A.; Holst C. R.; Huang P.; Kane B.; Kim M. H.; Li J.; Markby D.; Mohan M.; Noson K.; Plonowski A.; Richards S. J.; Robertson S.; Shaw K.; Stott G.; Stout T. J.; Young J.; Yu P.; Zaharia C. A.; Zhang W.; Zhou P.; Nuss J. M.; Xu W.; Kearney P. C. (2012) Discovery of XL413, a potent and selective CDC7 inhibitor. Bioorg. Med. Chem. Lett. 22, 3727–3731. 10.1016/j.bmcl.2012.04.024. [DOI] [PubMed] [Google Scholar]
- Sasi N. K.; Tiwari K.; Soon F.-F.; Bonte D.; Wang T.; Melcher K.; Xu H. E.; Weinreich M. (2014) The potent Cdc7-Dbf4 (DDK) kinase inhibitor XL413 has limited activity in many cancer cell lines and discovery of potential new DDK inhibitor scaffolds. PLoS One 9, e113300. 10.1371/journal.pone.0113300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A.; Valsasina B.; Croci V.; Menichincheri M.; Rainoldi S.; Marchesi V.; Tibolla M.; Tenca P.; Brotherton D.; Albanese C.; Patton V.; Alzani R.; Ciavolella A.; Sola F.; Molinari A.; Volpi D.; Avanzi N.; Fiorentini F.; Cattoni M.; Healy S.; Ballinari D.; Pesenti E.; Isacchi A.; Moll J.; Bensimon A.; Vanotti E.; Santocanale C. (2008) A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem. Biol. 4, 357–365. 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- Huggett M. T.; Tudzarova S.; Proctor I.; Loddo M.; Keane M. G.; Stoeber K.; Williams G. H.; Pereira S. P. (2016) Cdc7 is a potent anti-cancer target in pancreatic cancer due to abrogation of the DNA origin activation checkpoint. Oncotarget 7, 18495–507. 10.18632/oncotarget.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque A. A.; Fanous A. A.; Poh A.; Eastman A. (2008) Defective p53 signaling in p53 wild-type tumors attenuates p21waf1 induction and cyclin B repression rendering them sensitive to Chk1 inhibitors that abrogate DNA damage-induced S and G2 arrest. Mol. Cancer Ther. 7, 252–262. 10.1158/1535-7163.MCT-07-2066. [DOI] [PubMed] [Google Scholar]
- Wright G.; Golubeva V.; Remsing Rix L. L.; Berndt N.; Luo Y.; Ward G. A.; Gray J. E.; Schonbrunn E.; Lawrence H. R.; Monteiro A. N. A.; Rix U. (2017) Dual Targeting of WEE1 and PLK1 by AZD1775 Elicits Single Agent Cellular Anticancer Activity. ACS Chem. Biol. 12, 1883–1892. 10.1021/acschembio.7b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheghiani L.; Loew D.; Lombard B.; Mansfeld J.; Gavet O. (2017) PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep. 19, 2060–2073. 10.1016/j.celrep.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Lens S. M. A.; Voest E. E.; Medema R. H. (2010) Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer 10, 825–841. 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Castedo M.; Perfettini J.-L.; Roumier T.; Andreau K.; Medema R.; Kroemer G. (2004) Cell death by mitotic catastrophe: a molecular definition. Oncogene 23, 2825–2837. 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- Al-Ahmadie H.; Iyer G.; Hohl M.; Asthana S.; Inagaki A.; Schultz N.; Hanrahan A. J.; Scott S. N.; Brannon A. R.; McDermott G. C.; Pirun M.; Ostrovnaya I.; Kim P.; Socci N. D.; Viale A.; Schwartz G. K.; Reuter V.; Bochner B. H.; Rosenberg J. E.; Bajorin D. F.; Berger M. F.; Petrini J. H. J.; Solit D. B.; Taylor B. S. (2014) Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discovery 4, 1014–1021. 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitov M. I.; Greaser M. L.; Campbell K. S. (2009) GelBandFitter - A computer program for analysis of closely spaced electrophoretic and immunoblotted bands. Electrophoresis 30, 848–851. 10.1002/elps.200800583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J.; Otto W. R. (1992) Fluorimetric DNA assay for cell growth estimation. Anal. Biochem. 207, 186–92. 10.1016/0003-2697(92)90521-8. [DOI] [PubMed] [Google Scholar]
- Montano R.; Khan N.; Hou H.; Seigne J.; Ernstoff M.; Lewis L.; Eastman A. (2017) Cell cycle perturbation induced by gemcitabine in human tumor cells in cell culture, xenografts and bladder cancer patients:implications for clinical trial designs combining gemcitabine with a Chk1 inhibitor. Oncotarget 8, 67754–67768. 10.18632/oncotarget.18834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.