Abstract
Porcine deltacoronavirus (PDCoV) is a recently identified coronavirus that causes intestinal diseases in neonatal piglets with diarrhea, vomiting, dehydration, and post-infection mortality of 50–100%. Currently, there are no effective treatments or vaccines available to control PDCoV. To study the potential of RNA interference (RNAi) as a strategy against PDCoV infection, two short hairpin RNA (shRNA)-expressing plasmids (pGenesil-M and pGenesil-N) that targeted the M and N genes of PDCoV were constructed and transfected separately into swine testicular (ST) cells, which were then infected with PDCoV strain HB-BD. The potential of the plasmids to inhibit PDCoV replication was evaluated by cytopathic effect, virus titers, and real-time quantitative RT-PCR assay. The cytopathogenicity assays demonstrated that pGenesil-M and pGenesil-N protected ST cells against pathological changes with high specificity and efficacy. The 50% tissue culture infective dose showed that the PDCoV titers in ST cells treated with pGenesil-M and pGenesil-N were reduced 13.2- and 32.4-fold, respectively. Real-time quantitative RT-PCR also confirmed that the amount of viral RNA in cell cultures pre-transfected with pGenesil-M and pGenesil-N was reduced by 45.8 and 56.1%, respectively. This is believed to be the first report to show that shRNAs targeting the M and N genes of PDCoV exert antiviral effects in vitro, which suggests that RNAi is a promising new strategy against PDCoV infection.
Keywords: Porcine deltacoronavirus, RNA interference, Short hairpin RNA, Nucleocapsidgene, Swine testicular cells
Introduction
Porcine deltacoronavirus (PDCoV) is a new member of the Deltacoronavirus genus Coronavirus that causes intestinal disease. Clinical symptoms include vomiting, diarrhea, dehydration, and even death of piglets [1–3]. Since the first report of PDCoV in Hong Kong in 2012 [4] and the outbreak of PDCoV in the USA in 2014 [5], the novel porcine coronavirus has been detected in Canada, South Korea, Thailand, Mexico, and China [3, 6–8]. PDCoV has become an important pathogen affecting healthy development in the pig industry. However, there are currently no vaccines or treatments that can effectively control PDCoV [6].
PDCoV is an enveloped, single-stranded, and positive-sense RNA virus. The entire genome contains about 25,400 nucleotides [4]. PDCoV has four major structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) [1, 2, 9]. The detailed function of each PDCoV proteins is unknown. According to studies on other coronaviruses, M protein is the most abundant component of the viral envelope. It plays an important role in viral assembly process and budding [10, 11]. The M protein can also induce production of protective antibodies [10, 12]. The N protein of the coronavirus forms a helical nucleocapsid with genomic RNA and protects the viral genome from external interference [13, 14].
RNA interference (RNAi) is a process that effectively silences or inhibits the expression of a gene of interest, which is achieved by double-stranded RNA (dsRNA), which selectively inactivates the mRNA of the target gene. RNAi has been successfully used in infection inhibition studies of animal viruses, such as porcine epidemic diarrhea virus (PEDV) [15], influenza virus A [16], porcine transmissible gastroenteritis virus (TGEV) [17], and porcine reproductive and respiratory syndrome virus [18]. However, whether RNAi inhibits the replication of PDCoV has not been reported.
RNAi interferes with viral replication through short hairpin RNAs (shRNAs) or small interfering RNAs (siRNAs). shRNA is more stable, long-lasting, and more efficient than siRNA. Therefore, in this study, we constructed two shRNAs (pGenesil-M and pGenesil-N) in a plasmid expression system that targeted the M and N genes of PDCoV, and investigated the efficiency of shRNA-mediated RNAi of PDCoV replication in vitro.
Materials and methods
Cell culture, virus propagation, and titration
ST cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum (Zhejiang Tianhang Biotechnology Co. Ltd., Hangzhou, China) and 1% penicillin–streptomycin solution, and incubated in a 37 °C environment containing 5% carbon dioxide. The PDCoV strain HB-BD (GenBank No. MF948005) was propagated in ST cells as previously described [1]. After 80% of the cells developed a cytopathic effect (CPE) of viral infection, the culture (cells plus medium) was collected and subjected to three freeze–thaw cycles to lyse the cells. The virus titer was determined by the 50% tissue culture infectious dose (TCID50) as described previously [6].
shRNA sequence selection and shRNA expression plasmid construction
The predicted and analyzed shRNAs were obtained by predicting the M and N gene shRNAs of PDCoV using RNAi target finder software (www.Ambition.com/techlib/misc/siRNA). Using the BLAST program, the candidate shRNA sequences were aligned with the pig genome and other PDCoV sequences submitted to GenBank, and the specific strong shRNAs were selected. To ensure a similar RNAi effect on different PDCoV strains, two theoretically effective sequences at nucleotide positions 328–346 (M) and 270–288 (N) were selected. dsDNA sequences encoding the shRNAs were synthesized, with 4-nt 5′ single-stranded overhangs complementary to BamHI and HindIII-cleaved DNA at the ends. Scrambled siRNA sequences were designed as negative controls (NC). The sequences are shown in Table 1. Each ds-shRNA-coding sequence was ligated into the BamHI and HindIII restriction sites of the shRNA expression vector pGenesil-1 (Wuhan Genesil Biotechnology, China) and transformed into Escherichia coli competent cells. The recombinant plasmids were named pGenesil-M, pGenesil-N, and pGenesil-NC.
Table 1.
Sequences of designed shRNAs
| Plasmids | ShRNA |
|---|---|
| pGenesil-M | Sense:5′-GATCCGCAGGGATTATGGATCCAATTCAAGAGATTGGATCCATAATCCCTGCTTTTTTA-3′ |
| Antisense:5′-AGCTTAAAAAAGCAGGGATTATGGATCCAATCTCTTGAATTGGATCCATAATCCCTGCG-3′ | |
| pGenesil-N | Sense:5′-GATCCGCAACCACTCGTGTTACTTTTCAAGAGAAAGTAACACGAGTGGTTGCTTTTTTA-3′ |
| Antisense:5′-AGCTTAAAAAAGCAACCACTCGTGTTACTTTCTCTTGAAAAGTAACACGAGTGGTTGCG-3′ | |
| pGenesil-NC | Sense:5′-GATCCGCTATGTCAGTACTGCATTTTCAAGAGAAATGCAGTACTGACATAGCTTTTTTA-3′ |
| Antisense:5′-AGCTTTAAAAAAGCTATGTCAGTACTGCATTTCTCTTGAAAATGCAGTACTGACATAGCG-3′ |
The underlined sequences are siRNAs targeting the M and N genes and the bold italic letters indicate the loop sequence
Transfection of shRNA plasmids and virus infection
ST cells were seeded in a 24-well plate at 2 × 105 per well in 500 μL medium without antibiotics in a conventional manner and cultured for 18–24 h at 37 °C in a 5% CO2 environment. When the degree of monolayer cell confluence reached 70–80%, transfection was started according to the manufacturer’s instructions of LipoGene™ 2000 plus transfection reagent (US Everbright lnc., San Francisco, US). Briefly, 2 μg of transfection reagent was diluted with 50 μL of Opti-MEM and stand for 5 min at room temperature, then mixed with 50 μL diluted shRNA-expressing plasmid (1 μg shRNA-expressing plasmid was diluted with 50 μL of Opti-MEM). After 20 min, the cells of each well were washed three times and overlaid with 100 μL of the transfection mixture. The cells were incubated for 4 h at 37 °C, and the medium was changed. After 24 h, the cells were infected with 100 TCID50 PDCoV. ST cells infected with PDCoV but had undergone the transfection procedure without plasmid DNA added served as mock-transfected controls. Expression vector pGenesil-1 encodes the enhanced green fluorescent protein (EGFP). Therefore, EGFP expression in a cell line can be used as a transfection marker to evaluate the transfection efficiency. In this study, we used inverted fluorescence microscopy to capture images to evaluate the cell transfection efficiency and CPE.
Assay and determination of TCID50
PDCoV cultures in shRNA expression plasmid-transfected ST cells were harvested 48 h after virus infection. After three repeated freeze–thaw cycles, the virus was diluted from 10−1 to 10−10 and added to a 96-well plate with eight wells per dilution of virus. CPE was observed and recorded daily, and viral titer was measured by TCID50 using the Reed–Muench method as previously described [1, 6].
Analysis of RNA levels by real-time quantitative RT-PCR
N gene of PDCoV was used as a standard to assess the inhibitory effect of shRNA on viral replication. Total RNA was extracted from shRNA expression plasmids -transfected ST cells after 48 h of infection, and reverse transcription was performed using the PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa Biotechnology, Dalian, China). Quantitative real-time PCR (qRT-PCR) analysis was performed to amplify N gene using the cDNA as the template and the β-actin cDNA as the internal standard. The primers used are shown in Table 2. The quantitative RT-PCR contained 7.5 μL SYBR Mixture (CWBIO, Beijing, China), 0.5 μL (10 μM) forward primer, 0.5μL (10 μM) reverse primer, 1.5 μL cDNA template, and 10 μL RNase-free deionized distilled water. The reaction protocol was as follows: 95 °C for 600 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s; the melting curve stage comprised 95 °C for 10 s, 65 °C for 60 s, and 97 °C for 1 s, as well as 37 °C for 30 s. Each experiment was repeated three times. The relative number of viral RNA copies in cells interfered with different plasmids was calculated by the 2−ΔΔCt method.
Table 2.
Primers for real-time quantitative RT-PCR
| Primer | Sequences | Product length |
|---|---|---|
| PDCoV-NF | 5′- TACTGGTGCCAATGTCGGC -3′ | 262 bp |
| PDCoV-NR | 5′- AGTTGGTTTGGTGGGTGGC -3′ | |
| β-actin F | 5′- CCATCGTCCACCGCAAAT -3′ | 112 bp |
| β-actin R | 5′-CCAAATAAAGCATGCCAATC -3′ |
Results
Transfection efficiency of shRNA expression plasmids
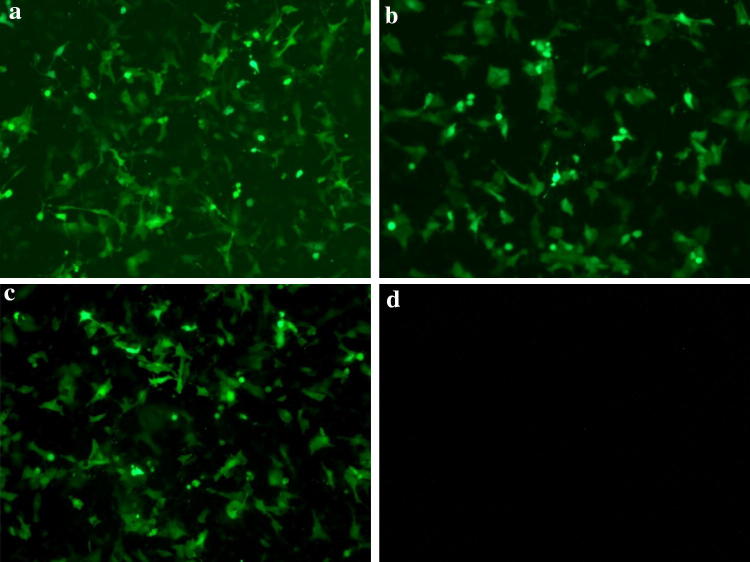
ST cells transfected with shRNA recombinant plasmids (pGenesil-M and pGenesil-N) and a scrambled shRNA recombinant plasmid (pGenesil-NC) were observed under fluorescence microscopy (Fig. 1). Normal ST cells showed no fluorescence, while ST cells transfected with recombinant plasmid showed fluorescence. The shRNA recombinant plasmids were successfully transfected into ST cells, and the transfection efficiency of the two recombinant plasmids and the scrambled shRNA plasmid was similar.
Fig. 1.
Fluorescence observation of ST cells at 24 h post-transfection. ST cells transfected with shRNA recombinant plasmid (pGenesil-M or pGenesil-N) and a scrambled shRNA recombinant plasmid (pGenesil-NC) at 24 h, examined by fluorescence microscopy. a pGenesil-M; b pGenesil-N; c pGenesil-NC; d mock-transfected ST cells
Examination of shRNA effects by CPE analysis
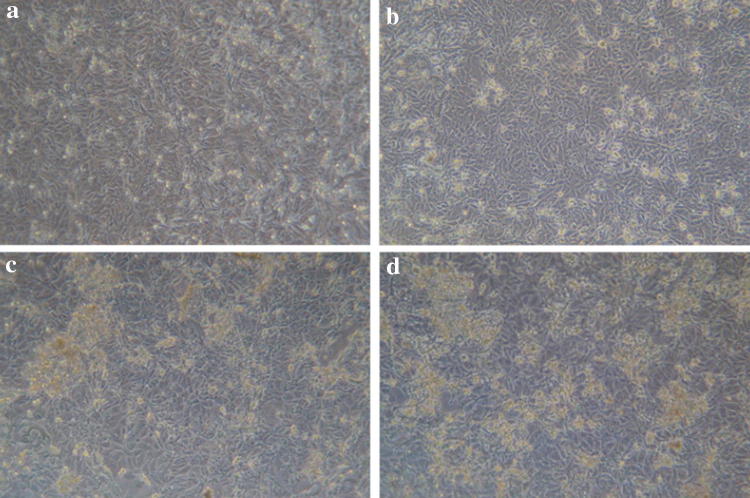
To analyze whether shRNA can prevent ST cells from exhibiting CPE due to PDCoV infection, recombinant plasmids pGenesil-M and pGenesil-N were transfected into ST cells seeded in triplicate in 24-well plates. The vector pGenesil-NC expressing the non-specific shRNA was used as a negative control. At 24 h after transfection, cells were infected with PDCoV at 100 TCID50, and cells were examined for CPE every day. CPE was observed 48 h after infection and photographed (Fig. 2). ST cells infected with virus only or ST cells transfected with a negative control plasmid (pGenesil-NC) became enlarged, round, dense granular cells, occurring individually or in clusters. We also observed signs of cell shrinkage and detachment from the monolayer. As for the cells transfected with plasmids pGenesil-N and pGenesil-M expressing specific shRNAs, observation showed that the extent of CPE was reduced.
Fig. 2.
CPE of ST cells 48 h after PDCoV infection (× 200). ST cells were transfected with different shRNA expression plasmids and infected with PDCoV at 100TCID50. a ST cells were pre-transfected with pGenesil-M; b ST cells were pre-transfected with pGenesil-N; c ST cells were pre-transfected with pGenesil-NC; d mock-transfected ST cells
Examination of shRNA effects on PDCoV replication by TCID50 assay
To analyze the inhibition of PDCoV replication by shRNA, viral titers in ST cells were calculated by the Reed–Muench method at 48 h after viral infection (Fig. 3). The results showed that the titers of PDCoV were 104.15 and 103.76 TCID50/mL in cells harboring pGenesil-M and pGenesil-N, respectively. The PDCoV titer was 105.35 TCID50/mL in cells pre-transfected with pGenesil-NC, which was similar to the titer of 105.27 TCID50/mL in mock-transfected cells. In comparison with the titers of virus produced by cells transfected with pGenesil-NC, that of pGenesil-M- and pGenesil-N-transfected cells corresponded to 13.2- and 32.4-fold reductions, respectively. The titers of virus produced by cells transfected with the recombinant plasmids pGenesil-N and pGenesil-M were significantly different from the virus titers transfected with pGenesil-NC (P < 0.05), while the difference between pGenesil-NC and mock-transfected cells was not significant. pGenesil-N showed higher inhibition efficiency than that of pGenesil-M shRNA.
Fig. 3.
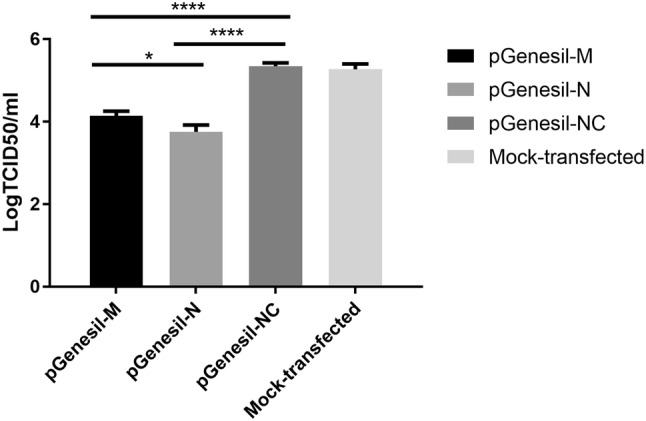
shRNA constructs interfere with PDCoV production in ST cells. TCID50 was calculated by the Reed–Muench method 48 h after infection. Virus yields were measured from TCID50. The data shown represent the means of three experiments. Error bars show the SD either ****P < 0.0001 pGenesil-M and pGenesil-N each compared with the negative control. *P < 0.05 pGenesil-M compared to pGenesil-N
Examination of shRNA inhibitory effect by real-time quantitative RT-PCR analysis
If RNAi is successful, replication of PDCoV is inhibited, and the amount of the corresponding M and N genes is less. We used the N gene as a standard to analyze the effect of shRNA inhibition of PDCoV replication. Real-time quantitative RT-PCR analysis of N gene level was normalized to the corresponding β-actin in the same sample (Fig. 4). The relative amount of N gene in mock cells was regarded as 1.000, whereas the relative amounts of N gene in cells infected with PDCoV after being transfected with pGenesil-M, pGenesil-N, and pGenesil-NC were 0.542, 0.439, and 1.079, respectively. Analysis of these data revealed that the amount of viral RNA in samples transfected with pGenesil-M and pGenesil-N was reduced by 45.8 and 56.1%, respectively, compared to the mock control. This suggests potent inhibition of PDCoV replication triggered by sequence-specific shRNAs in ST cells.
Fig. 4.
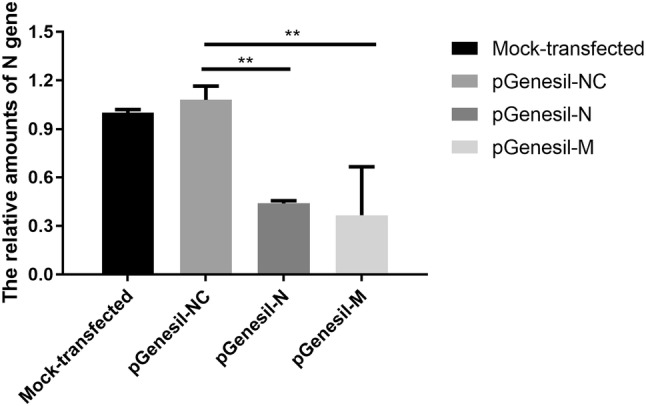
Inhibition of PDCoV replication by shRNAs in ST cells. Real-time quantitative RT-PCR detection of viral RNA copies relative to β-actin transcripts in the same sample. Data presented are the means of three repeat experiments. Error bars show SD. **P < 0.01 compared with the negative control
Discussion
PDCoV is a recently discovered porcine enteropathogenic coronavirus [8, 19–21]. Since 2015, PDCoV has emerged in many provinces, leading to significant economic losses in swine husbandry in China. However, there are presently no effective treatments or vaccines available to control PDCoV [6]. So, there is an urgency to develop an effective method for treatment of PDCoV.
RNAi is a gene silencing mechanism at the post-transcriptional level with high specificity and can inhibit gene expression with high efficiency. Therefore, RNAi has been considered as an effective strategy to protect against bacterial and viral pathogens [22, 23]. RNAi is triggered by endogenous or exogenous 21–23 nt RNA duplexes [17], and shRNA and siRNA are two commonly used RNA molecules to block gene expression [17, 24]. Compared with siRNA, the interference efficiency induced by shRNA was more effective. Recently, the interference efficiency of shRNA in some coronaviruses has been studied. Wang designed and constructed three recombinant plasmids targeting the M gene of porcine TGEV. After transfection into PK15 cells, M gene expression was reduced by 13, 68, and 70% [17]. Shen et al. [15] have constructed five shRNA-expressing plasmids targeting the N, M, and S genes of PEDV. PEDV RNA in Vero cells pre-transfected with these plasmids was reduced by 22–94.5%. However, because the success rate of PDCoV isolation was low [1, 6, 25], there are no reports on the use of shRNA to inhibit replication of PDCoV, which belongs to the same virus family as PEDV and TGEV.
In our previous study, PDCoV strain HB-BD was successfully isolated and serially passaged in cell culture and characterized. In this study, we designed two shRNAs based on the theoretically valid sequences of the M and N genes of PDCoV with nucleotide positions of 328–346 (M) and 270–288 (N), and established three shRNA recombinant expression plasmids to study whether shRNA-mediated RNAi inhibited PDCoV replication in vitro. To guarantee a similar RNAi effect on different PDCoV strains, the two theoretically effective sequences were analyzed by BLAST to ensure that they did not have any similar sequences in the swine genome, but shared 100% similarity with the published sequences of different PDCoV strains. Both the shRNAs inhibited PDCoV replication in ST cells. The interference properties were revealed by reductions in CPE formation, virus TCID50 titers, and viral RNA copy numbers in the infected cells. The CPE and TCID50 assay of pGenesil-M- and pGenesil-N-transfected cells showed viral suppression 48 h after infection and the titers of pGenesil-M- and pGenesil-N-transfected cells were reduced 13.2- and 32.4-fold, respectively. The real-time quantitative RT-PCR assay showed that the viral RNA copy number was reduced by 45.8% in pGenesil-M-transfected cells and 56.1% in pGenesil-M-transfected cells. The inhibitory efficiency of pGenesil-N was higher than that of pGenesil-M. Although the interference efficiency of shRNAs against PDCoV in our study was lower than that of the shRNAs targeting other coronaviruses [15, 17], these results indicate that RNAi against PDCoV mediated by shRNAs can inhibit PDCoV replication in vitro. A disadvantage of this transient transfection/expression system is that virus replication can be inhibited only in cells that are expressing the shRNA, and cells not expressing the shRNA can be infected. Therefore, one explanation for the lower interference efficiency of shRNAs against PDCoV in this study compared to shRNAs targeting other coronaviruses in other studies could be lower transfection efficiency of the shRNA expression plasmids.
In conclusion, our results indicate that both shRNAs plasmids targeting the M (328–346) and N (210–288) genes of PDCoV genome inhibit PDCoV replication in ST cells with high efficiency. Therefore, the two nucleotide positions are two potential targets for the inhibition of PDCoV replication by RNAi in vitro. However, whether the two shRNAs can inhibit PDCoV replication in vivo, and whether shRNAs targeting other nucleotide positions of the M and N genes or other genes also inhibit PDCoV replication need further research.
Acknowledgements
This research was supported by the Program of the Modern Agriculture Industry Technology System Foundation of Hebei Province (HBCT2018110207), Science and Technology Innovation Program of Hebei Province for graduate students (CXZZBS2019095), and Key R & D projects in Hebei Province, China (19226622D).
Author contributions
WYG performed the main experiments and wrote the paper; YL participated in editing the paper; BJL was involved in executing the study and editing the paper; YZZ and JHF participated in experimental design; JW, GFY, and SJC revised the paper.
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflicting interests.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wen-yuan Gu, Yan Li and Bao-jing Liu have contributed equally to this work.
Contributor Information
Yu-Zhu Zuo, Phone: + 86-312-7528581, Email: zuoyuzhu@163.com.
Jing-Hui Fan, Phone: + 86-312-7528581, Email: jinghui76@163.com.
References
- 1.Liu BJ, Zuo YZ, Gu WY, Luo SX, Shi QK, Hou LS, Zhong F, Fan JH. Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province China. Transbound Emerg Dis. 2018;65(3):874–882. doi: 10.1111/tbed.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014 doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA. Emerg Infect Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. 2015;53(5):1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, Zhang H, Lv DH, Li F, Wang D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Lee C. Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014 doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Chen J, Shi H, Chen X, Shi D, Feng L, Yang B. Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus. Virol J. 2012;9:225. doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan JH, Zuo YZ, Li JH, Pei LH. Heterogeneity in membrane protein genes of porcine epidemic diarrhea viruses isolated in China. Virus Genes. 2012;45(1):113–117. doi: 10.1007/s11262-012-0755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, Lin CH, Huang TH. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molenkamp R, Spaan WJ. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H, Zhang C, Guo P, Liu Z, Zhang J. Effective inhibition of porcine epidemic diarrhea virus by RNA interference in vitro. Virus Genes. 2015;51(2):252–259. doi: 10.1007/s11262-015-1242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui HY, Zhao GY, Huang JD, Jin DY, Yuen KY, Zheng BJ. Small interfering RNA targeting M2 gene induces effective and long term inhibition of influenza A virus replication. PLoS ONE. 2009;4(5):e5671. doi: 10.1371/journal.pone.0005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Dai X, Song H, Yuan P, Yang Z, Dong W, Song Z. Inhibition of porcine transmissible gastroenteritis virus infection in porcine kidney cells using short hairpin RNAs targeting the membrane gene. Virus Genes. 2017;53(2):226–232. doi: 10.1007/s11262-016-1409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou F, Liang S, Chen AH, Singh CO, Bhaskar R, Niu YS, Miao YG. A transgenic Marc-145 cell line of piggyBac transposon-derived targeting shRNA interference against porcine reproductive and respiratory syndrome virus. Vet Res Commun. 2012;36(2):99–105. doi: 10.1007/s11259-012-9519-9. [DOI] [PubMed] [Google Scholar]
- 19.Homwong N, Jarvis MC, Lam HC, Diaz A, Rovira A, Nelson M, Marthaler D. Characterization and evolution of porcine deltacoronavirus in the United States. Prev Vet Med. 2016;123:168–174. doi: 10.1016/j.prevetmed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madapong A, Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Srijangwad A, Tripipat T, Wegner M, Nilubol D. Complete genome sequence of porcine deltacoronavirus isolated in Thailand in 2015. Genome Announc. 2016 doi: 10.1128/genomeA.00408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouad AM, Soliman H, Abdallah ESH, Ibrahim S, El-Matbouli M, Elkamel AA. In-vitro inhibition of spring viremia of carp virus replication by RNA interference targeting the RNA-dependent RNA polymerase gene. J Virol Methods. 2018;263:14–19. doi: 10.1016/j.jviromet.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Man DK, Chow MY, Casettari L, Gonzalez-Juarrero M, Lam JK. Potential and development of inhaled RNAi therapeutics for the treatment of pulmonary tuberculosis. Adv Drug Deliv Rev. 2016;102:21–32. doi: 10.1016/j.addr.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]