Abstract
A maximum clade credibility tree constructed using the full-length spike (S) and hemagglutinin-esterase genes revealed that Vietnamese Bovine coronavirus (BCoV) strains belong to a single cluster (C1); therefore, they might share a common origin with Cuban and Chinese BCoV strains. The omega values of cluster 1 (C1) and cluster 2 (C2) were 0.15734 and 0.11613, respectively, and naive empirical bayes analysis identified two amino acid positions (179 and 501) in the S protein in C1 and three amino acid positions (113, 501, and 525) in that of C2 that underwent positive selection (p > 99%). The evolutionary rate of C1 was estimated to be 7.6206 × 10−4 substitutions/site/year, and the most recent common ancestor (tMRCA) of Vietnamese BCoVs was estimated to date back to 1962 (95% HPD 1950–1973). The effective population sizes of C1 and C2 underwent a rapid reduction after 2000 and 2004, respectively.
Electronic supplementary material
The online version of this article (10.1007/s11262-019-01647-1) contains supplementary material, which is available to authorized users.
Keywords: Bovine coronavirus, tMRCA, Evolutionary rate, Positive selection
Bovine coronavirus (BCoV) was first identified during an outbreak of diarrhea among neonatal calves in the 1970s [1]. Later, it occurred in association with winter dysentery in adult cattle and with respiratory tract disorders in calves and cattle [2, 3]. BCoV infection causes acute diarrhea in young calves and reduces milk production by dairy cows, both of which lead to significant economic losses [4]. More recently, preventing BCoV infection has become more important due to reported interspecies transmission of BCoV among feedlot cattle and zoo ruminants [5].
BCoV is a betacoronavirus (order Nidovirales, family Coronaviridae) that harbors a single-stranded, non-segmented, positive-sense RNA genome (26–31 kb) encoding five major structural proteins: the nucleocapsid (N) protein, the transmembrane (M) glycoprotein, the spike (S) glycoprotein, the envelope (E) protein, and the hemagglutinin-esterase (HE) glycoprotein [6]. The S glycoprotein comprises two subunits: S1 and S2. The N-terminal S1 subunit is a domain necessary for host surface receptor interaction prior to virus entry and is important for pathogenesis [7]. The HE protein also plays a role in virus entry and release from infected host cells [8]. The hypervariable region of the S protein is used to determine the genetic variability and evolution of BCoV virus [9, 10].
To date, BCoV infection has been reported in calves and adult cattle in many cattle-producing countries. In Vietnam, it has been reported that ELISA was applied to investigate the calf diarrhea in the Hue city and resulted that Bovine rotavirus and BCoV-positives of samples were 37.7% and 33.3%, respectively [11]. However, limited information is available about BCoV circulating in Vietnamese cow farms. Here, we examined the prevalence of BCoV infection among calves in Vietnam and genetically characterized Vietnamese BCoVs through phylogenetic and evolutionary analyses of the S and HE proteins.
Between 2017 and 2018, diarrheic fecal samples (n = 232) were collected from cows on farms located in Vietnam [Nam Dinh (ND), Phu Ly Ha Nam (PL), Duy Tien Ha Nam (DT), Binh Luc Ha Nam (BL), Moc Chau-Son La (MC), Vinh Phuc (VP), Nghe An (NA), Ha Tinh (HT), and TP HCM (HCM)]. All cows were aged between 0.5 and 72 months and had diarrhea. BCoV-positive feces were diagnosed using a Rapid BoviD-5 antigen kit (BioNote Inc., Hwaseong, Gyeonggi-do, Korea) and PCR analysis using previously described primer sets [4]. Sixteen of the fecal samples (from calf aged 0.5–4 months) tested positive for BCoV (6.9%, 16/232); the samples were designated ND65, PL83, PL84, DT97, BL104, MC199, VP200, NA226, HT293, HCM304, HCM305, HCM306, HCM307, HT315, HT316, and HT317 (Supplemental Table and Figure).
The full-length S and HE gene sequences of 16 Vietnamese BCoVs were aligned using Clustal X 1.83 software [12] and compared with reference sequences (100 spike genes and 41 HE genes) collected from the NCBI GenBank database (Figs. 1, 2a). The full-length S gene sequence of Vietnamese BCoV contained 4092 nucleotides, which encode 1363 amino acid (aa) residues. The amino acid sequence showed 98.1–99.4% with Cuba isolates from 2009 to 2011, 98.0–98.8% with Chinese strain (AKS01) from 2015, 97.6–99.2% homology with USA stain (ENT) from 1998, and 96.8–98.5% homology with European strains from 1992 to 2014.
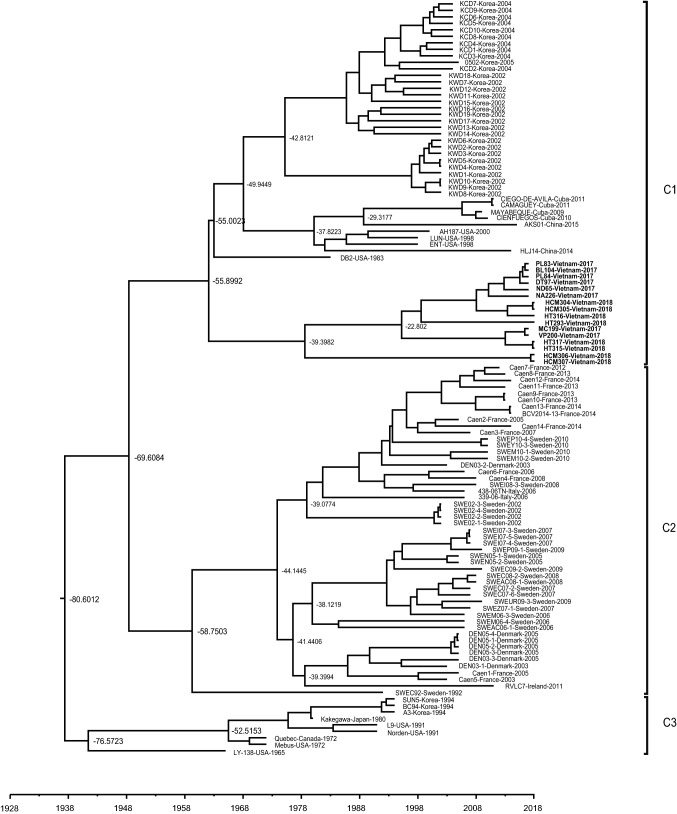
Fig. 1.
Bayesian phylogenetic tree (under a strict molecular clock) for the full-length S gene of BCoV. The maximum clade credibility (MCC) tree was built using the best model (GTR + I + G) and is scaled to time (horizontal axis). The times at which C1, C2, and C3 diverged from a common ancestor are denoted at the nodes
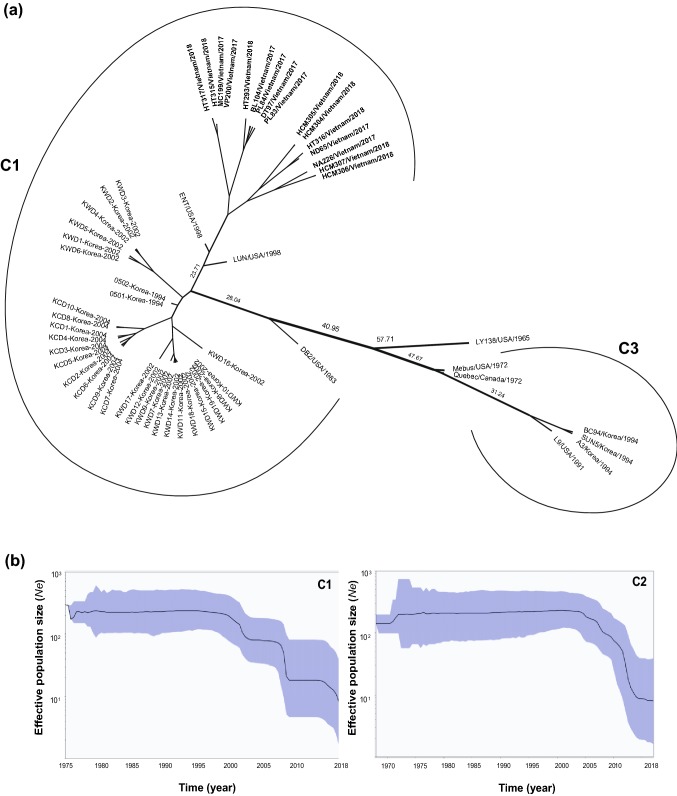
Fig. 2.
MCC tree constructed using the BEAST program and based on the full-length HE gene nucleotide sequences of BCoV (a), and a Bayesian skyline plot (BSP) based on the full-length S gene sequences of cluster 1 and 2 (b). The most probable year for the tMRCA within each lineage is shown (a). The plot depicts changes in the effective population size (Ne), and the dark line shows the effective population size estimated overtime (b). The upper and lower lines indicate the 95% HPD range of the BSP
The software jModelTest 2.1.10 is used to estimate the best-fit model using Akaike and Bayesian information criteria [13]. The best-fit model for the BCoV S and HE genes was the GTR + I + G model. BCoV sequences were used to generate a BEAST input file using BEAUti within BEAST package v1.8.1. The rates of nucleotide substitution per site/per year and the most recent common ancestor (tMRCA) were estimated using a Bayesian MCMC approach [14]. Each dataset was simulated using the following options: generation, 80,000,000; burn-in, 10%; and ESSs, > 300. The resulting convergence was analyzed using Tracer 1.5 [14]. Trees were presented as MCC trees using TreeAnnotator 1.7.4 and visualized using Figtree 1.4 [15]. The MCC tree for the S gene indicated that BCoV strains were divided into three diverse clusters (Fig. 1). Cluster 1 (C1) comprised 56 BCoVs isolates (identified between 1983 and 2018) derived from the USA (n = 4), China (n = 2), Korea (n = 30), Cuba (n = 4), and Vietnam (n = 16). Cluster 2 (C2) contained 51 BCoVs identified between 1992 and 2014 in European countries: Denmark (n = 7), Sweden (n = 26), France (n = 15), Ireland (n = 1), and Italy (n = 2). Cluster 3 (C3) comprised 9 BCoV strains identified between 1965 and 1994 in Canada (n = 1), USA (n = 4), Korea (n = 3), and Japan (n = 1). A previous study suggests that HCoV-OC43, BCoVs, and BCoV-like viruses are distributed within three main sub-clusters, named C1 (BCoVs and BCoV-like from America and Asia), C2 (BCoVs from Europe), and C3 (prototype, vaccine, or attenuated BCoV strains) [16]. Antigenic variability and evolution of BCoVs has been reported mainly in strains from USA and Korea, suggesting that BCoV strains take different evolutionary pathways in different countries [4]. Cuban BCoVs clustered with USA BCoV strains (the US/OH1/2003 strain derived from an antelope and the US/OH3/2003 strain derived from a giraffe), suggesting that these viruses have a common ancestor [17].
The evolutionary rate of BCoV was estimated to be 3.2013 × 10−4 substitutions/site/year (95% highest posterior density (HPD) 2.3252 × 10−4–4.1831 × 10−4) according to ESS (396.6217), and the tMRCA of 116 BCoVs was estimated to be 1937 (95% HPD 1922–1951: calendar). According to the MCC tree, C1 first appeared in 1962 (95% HPD 1950–1973: calendar), C2 in 1948 (95% HPD 1928–1964: calendar), and C3 in 1943 (95% HPD 1932–1954: calendar) (Table 1). The tMRCA of Vietnam strains was estimated to be 1962 (95% HPD 1950–1973: calendar). Molecular clock analysis of S gene sequences suggested that BCoV and CRCoV diverged from a common ancestor in 1951, whereas BCoV and HCoV-OC43 diverged in 1899 [5].
Table 1.
Mean estimations for the rate of evolution and tMRCAs and positively selected sites of different lineages for BCoV spike genes
| Lineage | Substitution rate (× 10−4 subs/site/year) | TMRCA (calendar year) | Omega value (dN/dS) | Positively selected sites (p > 95%; *p > 99%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% HPD lower | 95% HPD upper | Mean | 95% HPD lower | 95% HPD upper | NEB | BEB | ||
| BCoV | 3.2013 | 2.3252 | 4.1831 | 1937 | 1922 | 1951 | - | - | - |
| C1 | 7.6206 | 6.2199 | 9.1537 | 1962 | 1950 | 1973 | 0.15734 | 11, 179*, 499, 501*, 1352 | 11*, 179*,256, 412, 499*, 501*, 716, 1352* |
| C2 | 5.479 | 3.9751 | 6.9255 | 1948 | 1928 | 1964 | 0.11613 | 35, 113*, 501*, 525* | 23, 35*, 113*, 118, 257, 501*, 525*, 540, 590 |
| C3 | 69.803 | 1.2717 | 15.65 | 1943 | 1932 | 1954 | 1.90805 | 40*, 175, 179*, 253*, 484*, 906 | 40*, 175, 179*, 253*, 484, 905 |
| V | - | - | - | 1962 | 1950 | 1973 | 0.10297 | - | - |
V Vietnam BCoV strains, NEB naive empirical bayes analysis, BEB bayes empirical bayes analysis, - not test
*Positively selected sites identified with posterior probability > 99% level
To test the hypothesis that the full-length S gene of BCoV undergoes positive selection, we examined site models and branch site models implemented in the BASEML and CODEML program of the PAML v4.6 package [18]. The substitution rate ratios for non-synonymous (dN) versus synonymous (dS) mutations (ω) were calculated. The ω ratio for C1, C2, C3, and Vietnam BCoV strains were 0.15734, 0.11613, 1.90805, and 0.10297, respectively (Table 1). Naive empirical Bayes (NEB) analysis identified positively selected sites within the S protein in the C1 cluster as aa 179 and aa 501 (p > 99%), whereas Bayes empirical Bayes analysis identified four aa positions (11, 179, 499, 501, 1352) (p > 99%) (Table 1). NEB analysis estimated positively selected sites in the C2 cluster as aa 113, aa 501, and aa 525 (p > 99%) (Table 1). Two strong positive selection sites were detected within the receptor-binding subunit of the S protein gene, spanning aa residues 109–131 and aa 497–527 [5]. The selection pattern along the S glycoprotein implies adaptive evolution of BCoVs, suggesting a successful mechanism by which the virus continually circulates among cattle and other ruminants [5].
The BCoV HE gene separated into two clusters (Fig. 2a). C1 comprised 50 BCoVs from the USA (n = 3), Vietnam (n = 16), and Korea (n = 31), and C3 comprised 6 BCoVs from the USA (n = 2), Canada (n = 1), and Korea (n = 3). The evolutionary rate of the BCoV HE gene was estimated to be 4.5630 × 10−4 substitutions/site/year (95% HPD 3.1982 × 10−4–6.0408 × 10−4) according to ESS (4448.7229). The tMRCA on the MCC tree of BCoV HE gene suggests that C1 and C3 diverged 41.3604 years ago (95% HPD 35.6184–47.4545) and 63.4262 years ago (95% HPD 55.8045–71.4806), respectively (data not shown).
The effective population size for the BCoV C1 cluster estimated by Bayesian skyline analysis fell sharply at three separate times. The first fall was from 196.7854 (95% HPD 403.4424–72.53619) in 2000 to 92.57811 (95% HPD 199.8897–36.79424) in 2002. The second fall was from 82.83793 (95% HPD 176.1481–30.70227) in 2007 to 24.81323 (95% HPD 81.35125–4.641601) in 2008. Finally, the third fall was from 21.73334 (95% HPD 75.96825–3.794129) in 2015 to 11.40183 (95% HPD 38.80617–1.621968) in 2017 (Fig. 2b). The effective population size for the BCoV C2 cluster fell once, from 201.6739 (95% HPD 400.4581–74.48877) in 2004 to 13.38442 (95% HPD 47.3822–2.208484) in 2014 (Fig. 2b).
In conclusion, Vietnam BCoV sequences might share a common ancestor with the Cuban and Chinese BCoV strains [high nucleotide sequence similarity within the same cluster, (C1)]. Molecular clock analysis of S gene sequences estimated that the time of divergence from a common ancestor of C1 and C2 was estimated to be 1962. The effective population sizes for the BCoV C1 and C2 clusters fell rapidly from 2000 to 2004, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
DJA, JS, and DT conceived and designed the experiments. VPL, SC, BTN, and NTL performed the experiments. DT, RMC, GNP, and ISC analyzed the data. DJA and JS wrote the paper and designed the figure. All authors read and approved the final manuscript.
Funding
This study was funded by a grant (Project Code No. I-1543083-2017-19-0101) from the Animal and Plants Quarantine Agency (QIA), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea (2017).
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This article did not involve studies of human subjects or animals.
Footnotes
Edited by Zhen F. Fu.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jihye Shin and Dongseob Tark are Co-first authors.
References
- 1.Mebus CA, Stair EL, Rhodes MB, Twiehaus MJ. Neonatal calf diarrhea: propagation, attenuation, and characteristics of a coronavirus like agent. Am J Vet Res. 1974;34:145–150. [PubMed] [Google Scholar]
- 2.Cho KO, Hoet AE, Loerch SC, Wittum TE, Saif LJ. Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am J Vet Res. 2001;62:1436–1441. doi: 10.2460/ajvr.2001.62.1436. [DOI] [PubMed] [Google Scholar]
- 3.Chouljenko VN, Kousoulas KG, Lin X, Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. 1998;17:33–42. doi: 10.1023/A:1008048916808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SJ, Jeong C, Yoon SS, Choy HE, Saif LJ, Park SH, Kim YJ, Jeong JH, Park SI, Kim HH, Lee BJ, Cho HS, Kim SK, Kang MI, Cho KO. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J Clin Microbiol. 2006;44:3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidokhti MR, Traven M, Krishna NK, Munir M, Belak S, Alenius S, Cortey M. Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. J Gen Virol. 2013;94:2036–2049. doi: 10.1099/vir.0.054940-0. [DOI] [PubMed] [Google Scholar]
- 6.Lai MMC, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultze B, Gross HJ, Brossmer R, Herrler G. The S Protein of bovine coronavirus is a hemagglutinin recognizing 9-o-acetylated sialic acid as a receptor determinant. J Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultze B, Wahn K, Klenk HD, Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991;180:221–228. doi: 10.1016/0042-6822(91)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandão PE, Gregori F, Richtzenhain LJ, Rosales CAR, Villarreal LYB, JerezJ A. Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43. Arch Virol. 2006;151:1735–1748. doi: 10.1007/s00705-006-0752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasoksuz M, Sreevatsan S, Cho KO, Hoet AE, Saif LJ. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. 2002;84:101–109. doi: 10.1016/S0168-1702(02)00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chào NV, Hòa NX, Hải PV, Hùng PHS (2014) Application of ELISA method to determine the cause of calf diarrhea in the Hue city, Vietnam. Tạp chí khoa học, Đại học Huế 94:17–25 (Vietnamese)
- 12.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darriba D, Taboada GI, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Method. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond AJ, Suchard MA, Cie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambaut A. FigTree v112. Edinburgh: Institute of Evolutionary Biology, Univ Edinburgh; 2008. [Google Scholar]
- 16.Kin N, Miszczak F, Dianocourt L, Caro V, Moutou F, Vabert A, Ar Gouilh M. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect Genet Evol. 2016;40:186–191. doi: 10.1016/j.meegid.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez N, Brandao PE, de Souza SP, Barrera M, Santana N, de Arce HD, Perez LJ. Molecular and phylogenetic analysis of bovine coronavirus based on the spike glycoprotein gene. Infect Genet Evol. 2012;12:1870–1878. doi: 10.1016/j.meegid.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z. PMAL 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.