Abstract
A new family of IFNs called type III IFN or IFN-λ has been described, and shown to induce antiviral activity against several viruses in the cell culture. In this study, the molecular cloning, expression, and antiporcine epidemic diarrhea virus (PEDV) activity of porcine IFN-λ3 (poIFN-λ3) were reported. The full-length poIFN-λ3 cDNA sequence encoded 196 amino acids with a 23 amino acid signal peptide. Sequence alignments showed that poIFN-λ3 had an amino acid sequence similarity to Ovis aries (78.1 %), Bos taurus (76.0 %), Tupaia belangeri (71.3 %), Equus caballus (69.9 %), and Homo sapiens (69.9 %). The phylogenetic analysis based on the genomic sequences indicated that poIFN-λ3 is located in the same branch as B. taurus and O. aries IFN-λ3. The poIFN-λ3 without a signal anchor sequence was efficiently expressed in Escherichia coli, and the purified recombinant poIFN-λ3 exhibited significant antiviral effects against PEDV in a dose- and time-dependent manner. This inhibitory effect of poIFN-λ3 on PEDV was observed under three different treatment conditions. The highest inhibition of PEDV was observed in Vero E6 cell cultures pretreated with poIFN-λ3 (prior to PEDV infection). In addition, poIFN-λ3 was able to induce the expression of IFN-stimulated genes, including ISG15, OAS1, and Mx1 in Vero E6 cells. These data demonstrate that poIFN-λ3 has antiviral activity against PEDV and may serve as a useful biotherapeutic candidate to inhibit PEDV or other viruses in swine.
Keywords: Porcine IFN-λ3, Antiviral activity, PEDV
Interferons (IFNs) play an important role in the innate and adaptive immune responses against viral infections [20]. Type III IFNs (IFN-λs) were first identified in 2003 and are encoded by 1–3 functional genes in most mammalian species [22]. Humans and chickens have three IFN-λs as follows: IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 [12]. Mice have genes only for IFN-λ2 and IFN-λ3 [7]. Two members, IFN-λ1 (IL-29) and IFN-λ3 (IL-28B), have been recently identified in swine [21].
Distinct from the receptors for type I and II IFNs, type III IFN exerts its action through a heterodimeric cellular receptor that is composed of IL28-Rα, a type III IFN specific subunit, and IL10-Rβ, a subunit shared by other IL-10-related cytokines [18]. As IFN-λs bind to these unique receptors, they activate Janus kinase I, together with signaling transducers and activators of transcription (STAT) factors 1 and 2, resulting in the formation of the IFN-stimulated gene factor 3 (ISGF3) complexes [14]. This then induces the transcription of IFN-stimulated genes (ISGs), such as 2′,5′-oligoadenylate synthetase (2′,5′-OAS) and MxA genes [14]. IFN-λs share a number of common biological characteristics with type I IFN, such as antiviral, antiproliferative, and antitumor activities [3, 8, 9, 17]. The expression of IFN-λs can be induced by viral infection or treatment with poly (I:C) as well as lipopolysaccharide (LPS) [17]. Previous studies have demonstrated that IFN-λs can induce protection against several viruses in cell culture, such as West Nile virus [16], influenza A virus [10], porcine reproductive and respiratory syndrome virus [15], and foot-and-mouth disease virus [5].
Porcine epidemic diarrhea virus (PEDV) is a member of the family Coronaviridae, and is an important etiological agent of lethal watery diarrhea in piglets, resulting in large economic losses in many Asian and European countries [23, 24]. Previous research has demonstrated that PEDV infection suppresses the production of IFN-β, and viral papain-like protease 2 has been identified as a type I interferon antagonist [4, 6, 24, 25]. In this study, poIFN-λ3 was cloned and expressed in Escherichia coli using the pET-32a vector. In addition, the molecular characterization of poIFN-λ3 was analyzed, and the antiviral activity of poIFN-λ3 against PEDV was evaluated. Furthermore, a real-time quantitative PCR assay was used to quantify the expression levels of ISGs, OAS1, and Mx1 induced by poIFN-λ3 to elucidate the mechanism of poIFN-λ3 antiviral efficacy.
Based on our experiments, poIFN-λ3 gene was amplified using RT-PCR from ST cells stimulated with poly (I:C) and subcloned into pET-32a expression vector, transformed into host bacteria Rosetta gami B. The recombinant poIFN-λ3 was expressed by IPTG induction and existed in inclusion bodies (Fig. 1). PoIFN-λ3 was purified by Ni–NTA affinity chromatography using an ÄKTA purifier FPLC (GE Healthcare). After refolding, the eluted protein was dialyzed against phosphate-buffered saline (PBS) and filtered through a 0.22-μm filter and stored at −80 °C for further use. The purified poIFN-λ3 protein was tested via the CPE inhibition assay using Vesicular stomatitis virus (VSV)/MDBK systems. The results demonstrated that poIFN-λ3 possessed an anti-VSV activity.
Fig. 1.
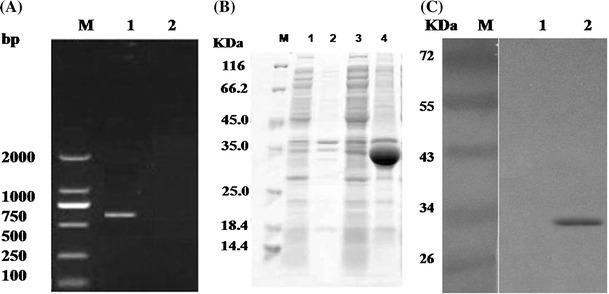
Cloning, expression, and purification of recombinant poIFN-λ3 in R. gami B. a Molecular cloning of poIFN-λ3. b SDS-PAGE analysis of samples. M, molecular weight marker; lane 1 the supernatant of bacterial cells harboring pET-poIFN-λ3 without IPTG induced; lane 2 the precipitation of bacterial cells harboring pET-poIFN-λ3 without IPTG induced; lane 3 the supernatant of bacterial cells harboring pET-poIFN-λ3 with 0.4 mM IPTG induced for 16 h at 20 °C; lane 4 the precipitation of bacterial cells harboring pET-poIFN-λ3 with 0.4 mM IPTG induced for 16 h at 20 °C. c Western blot using an anti-His antibody. M, molecular weight marker; lane 1 R. gami B extract transformed by pET-32a (+) (negative control); lane 2 R. gami B extract transformed by pET-poIFN-λ3
The sequencing results demonstrated that the full-length cDNA of poIFN-λ3 contains 588 bp and encodes 196 amino acid residues. The identified poIFN-λ3 sequence encodes a protein of MW = 21486.09 with a pI of 9.27 (http://web.expasy.org/compute_pi/). Moreover, a predicted signal peptide is located between amino acids 1 and 23 [2] and a putative N-linked glycosylation sequence was between amino acids 71 and 74 (http://www.cbs.dtu.dk/services/NetNGlyc/) (Fig. 2a, c).
Fig. 2.

Amino acid sequences analysis, phylogenetic tree, and prediction of a signal peptide for poIFN-λ3. a Alignment of amino acid sequences for porcine, B. taurus, O. aries, T. belangeri, Pan troglodytes, Equus caballus, Mus musculus, Gallus silurana, and Homo sapiens IFN-λ3. The numbers indicate the amino acid position. Predicted signal peptide: amino acids 1–23 (boxed in blue). Putative N-glycosylation site: amino acids 71–74 (boxed in red). b A phylogenetic tree of the nine identified or predicted IFN-λ3 from different species. The uprooted tree was built using the neighbor-joining method based on the alignment of IFN-λ3 amino acid sequences. The scale bar is 0.1. c The signal peptide of poIFN-λ3 was predicted by SignalP3.0 Server. C, S, and Y scores indicate cleavage sites, ‘signal peptide-ness,’ and combined cleavage site predictions, respectively (Color figure online)
To gain insight into the poIFN-λ3, BLASTN was performed using the IFN-λ3 sequences from nine species in GenBank. The nucleic acid sequence analysis revealed that poIFN-λ3 had a high similarity to Ovis aries IFN-λ3 (GenBank No. XM_004015683; 87.4 %), Bos taurus IFN-λ3 (GenBank No.XM_003585012; 85.5 %), Tupaia belangeri IFN-λ3 (GenBank No. JX185489; 80.9 %), Pan troglodytes IFN-λ3 (GenBank No. XM_001135462; 80.9 %), Homo sapiens IFN-λ3 (GenBank No. NM_172139; 78.9 %), and Equus caballus IFN-λ3 (GenBank No. NM_172139; 78.1 %), respectively. The amino acid sequence analysis results showed that poIFN-λ3 also has a high identity to O. aries (78.1 %), B. taurus (76.0 %), T. belangeri (71.3 %), E. caballus (69.9 %), and H. sapiens (69.9 %) IFN-λ3. The phylogenetic tree was constructed using the MEGA program and a neighbor-joining analysis method based on the DNA sequences. The phylogenetic analysis indicated that the poIFN-λ3 is located on the same branch as the B. taurus and O. aries genes (Fig. 2b).
For the studies of antiviral activity, PEDV strain CV777 was used as a model virus to evaluate the antiviral activity of poIFN-λ3. The virus was propagated and titrated in Vero E6 cells. Recombinant poIFN-λ3 were expressed, purified, and stored at −80 °C in our laboratory. For the anti-PEDV effects, Vero E6 cells were incubated with poIFN-λ3 either 24 h prior to PEDV infection, simultaneously with PEDV infection, or 12 h after PEDV infection. The productive PEDV infection was determined at day 2 postinfection. The viral titer was determined using TCID50. Also, the amounts of viral RNAs were detected by a real-time quantitative RT-PCR [13]. The results indicated that poIFN-λ3 could effectively inhibit the PEDV replication under three different treatment conditions. The highest inhibition of PEDV was observed in Vero E6 cell cultures pretreated with poIFN-λ3 (prior to PEDV infection) (Fig. 3a). To evaluate the dose-dependent effect of poIFN-λ3 on PEDV infection, Vero E6 cells were pretreated with poIFN-λ3 (10, 100, or 1000 ng/mL doses of poIFN-λ3) for 24 h prior to PEDV infection. The cells were then infected with PEDV for 2 h, washed, and replenished with fresh medium containing the indicated poIFN-λ3. Then, after 48 h infection, the cells were submitted to three freeze–thaw cycles and titrated by TCID50. Treatment with 10, 100, or 1000 ng/mL doses of poIFN-λ3 could protect Vero E6 cells from PEDV infection (Fig. 3c). As shown in Fig. 3b, treatment of Vero E6 cells with poIFN-λ3 (100 ng/mL) could inhibit the replication of PEDV at 12, 24, 36, and 48 h after infection. Therefore, poIFN-λ3 could significantly inhibit PEDV propagation in a dose- and time-dependent manner. In addition, the results demonstrated that the poIFN-λ3 can effectively reduce the amounts of PEDV RNAs in samples poIFN-λ3 prior to PEDV infection. And, accompanying dose of poIFN-λ3 increased (from 10 to 1000 ng/ml) and PEDV RNAs were reduced (Fig. 3d, e). These results were consistent with the TCID50 assay.
Fig. 3.
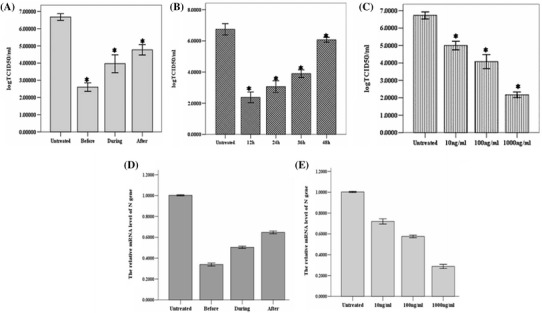
Effect of poIFN-λ3 on PEDV infection of Vero E6 cells. a Effect of poIFN-λ3 on PEDV infection and replication under three different conditions. Vero E6 cells were pretreated with poIFN-λ3 (100 ng/mL) for 24 h, and then infected with PEDV CV777 strain (Before) or coincubated with poIFN-λ3 (100 ng/mL) and PEDV CV777 strain at the same time (during), or 12 h after PEDV infection, Vero E6 cells were treated with 100 ng/mL poIFN-λ3 (After). b Time-dependent effect of poIFN-λ3 on PEDV infection. Vero E6 cells were infected with PEDV CV777 strain for 12 h and then treated with poIFN-λ3 at a concentration of 100 ng/mL. The PEDV titer in the supernatant at the indicated time point was titrated by TCID50. c Dose-dependent effect of poIFN-λ3 on PEDV infection. Vero E6 cells were infected with PEDV CV777 strain for 12 h and then treated with poIFN-λ3 at the indicated concentrations. The PEDV titer in the supernatant was analyzed at 48 h postinfection. d, e Real-time quantitative RT-PCR detection of PEDV-N mRNA transcripts relative to β-actin transcripts in the same sample. The mean of three repeat experiments performed in triplicate is shown and error bars represent the SD. d is under three different conditions and e is at different doses of poIFN-λ3
To better illustrate the mechanism of antiviral activity elicited by poIFN-λ3 in Vero E6 cells, for the analysis of gene expression, we focused on ISGs (including ISG15, Mx1, and OAS1 and the primers of these three genes are listed in Table 1) in which the expression has been shown to be up regulated by type III IFN in other species [1]. Vero E6 cells were treated with 100 ng/mL poIFN-λ3 and cell samples were collected 24 h posttreatment. Untreated Vero E6 cells were also collected as a calibrator to evaluate the mRNA levels of the chosen ISGs. GAPDH was used as a housekeeping gene for sample normalization. Total RNA was extracted from these Vero E6 cells, and qRT-PCR was performed using LightCycler® 96 SW 1.1 system and a SYBR Green I PCR master mix (Baoshengwu, Dalian, China). The samples were analyzed in triplicate. Levels of mRNA were calculated using the method, which expresses mRNA in treated cells relative to untreated cells after normalizing to GAPDH. As shown in Fig. 4, all tested IFNs induced a significant increase in the mRNA expression of the antiviral genes ISG15, OAS1, and Mx1, all of which have well-known antiviral properties and may affect PEDV replication.
Table 1.
Primer sequences used in this study
| Genes | Primer names | Sequence (5′–3′) | Size (bp) |
|---|---|---|---|
| OAS1 | OAS1-F | GGTTGTCTTCCTCAGTCCTC | 136 |
| OAS1-R | AGCCTGGACCTCAAACTTCA | ||
| Mx1 | Mx1-F | GCAGCCAGTACGAGGAGAAG | 153 |
| Mx1-R | CTCCTGACAGTGCCTCCAAC | ||
| ISG15 | ISG15-F | GGGCAACGAGTTCCAGGT | 193 |
| ISG15-R | CACCACCAGCAGGACCGT | ||
| GAPDH | GAPDH-F | AGCCAAAAGGGTCATCATCT | 180 |
| GAPDH-R | ATGAGTCCTTCCACGATACC | ||
| N | N-F | CGCAAAGACTGAACCCACTAATTT | |
| N-R | TTGCCTCTGTTGTTACTTG GAGAT | 198 | |
| β-actin | Actin-F | GGACTTCGAGCAGGAGATGG | |
| Actin-R | AGGAAGGAGGGCTGGAAGAG | 138 |
Fig. 4.
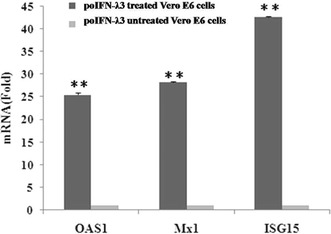
Expressions of OAS1, Mx1, and ISG15 induced by poIFN-λ3. The levels of OAS1, Mx1, and ISG15mRNA were measured by quantitative PCR, and the results were normalized by the GAPDH levels of each sample. Values represent the mean ± SD of three independent tests. P < 0.01 compared with the untreated Vero E6 cells
PEDV replicates in the small intestine epithelial cells, resulting in an acute and highly contagious enteric disease [11]. Recently, studies have proved the importance of IFN-λ for restricting viral infection in the intestinal epithelium [19]. IFN-λ plays a potent antiviral state by regulating the expression of IFN-stimulated genes (ISGs). Therefore, IFN-λ is important to efficiently restrict enteric coronavirus infections in the intestinal epithelial surface, including PEDV, TGEV, and possibly PDCoV infections. Correspondingly, these viruses may influence the host IFN-λ response. So, it is beneficial to understand the pathogenic mechanisms of porcine enteric coronaviruses and host responses to such infections [26].
In summary, poIFN-λ3 was successfully cloned, expressed, and possesses antiviral activity against PEDV in a dose- and time-dependent manner in vitro, suggesting that poIFN-λ3 may be a broad-spectrum antiviral agent. The treatment of Vero E6 cells with poIFN-λ3 induced the expressions of ISG15, OAS1, and Mx1. Moreover, this study provides a foundation to utilize type III IFNs as useful antiviral agents against swine infectious diseases.
Author contributions
All authors have reviewed the final version of the manuscript and approved it for publication.
Funding
This study was supported by the National Natural Science Foundation of China (No. 31302101), the Science and Technology Planning Project of Guangdong Province, China (Nos. 2014B070706011, 2014A010107022, and 2015A020209081), the International S&T Cooperation Program of China (No. 2014DFA31730), and the Special Fund for Agro-scientific Research in the Public Interest (201303046).
Compliance with ethical standards
Conflict of interest
All the authors have no conflict of interest to declare.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
All authors have reviewed the final version of the manuscript and approve it for publication.
Footnotes
Haiyan Shen and Chunhong Zhang have contributed equally to this study and are regarded as joint first authors.
References
- 1.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 3.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diebold J, Diepolder H, Adler B, Auernhammer CJ, Göke B. J. Dambacher. Am. J. Physiol. Gastrointest. Liver. Physiol. 2005;289:960–968. doi: 10.1152/ajpgi.00126.2005. [DOI] [PubMed] [Google Scholar]
- 4.Cao L, Ge X, Gao Y, Herrler G, Ren Y, Ren X, Li G. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Díaz-San Segundo F, Weiss M, Perez-Martín E, J.Koster M, Zhu J, Grubman MJ, de los Santos T. Virology. 2011;41:283–292. doi: 10.1016/j.virol.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly RP, Kotenko SV. J. Interf. Cytok. Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong SH, Cho O, Kim K, Shin HJ, Kotenko SV, Park S. Virus Res. 2007;126:245–249. doi: 10.1016/j.virusres.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, Ho WZ. J. Virol. 2009;83:3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichihashi T, Asano A, Usui T, Takeuchi T, Watanabe Y, Yamano Y. Vet. Immunol. Immunopathol. 2013;156:141–146. doi: 10.1016/j.vetimm.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Jung K, Saif LJ. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karpala AJ, Morris KR, Broadway MM, McWaters PGD, O’Neil TE, Goossens KE, Lowenthal JW, Bean AGD. J. Interf. Cytok. Res. 2008;28:341–350. doi: 10.1089/jir.2007.0117. [DOI] [PubMed] [Google Scholar]
- 13.Kim SH, Kim IJ, Pyo HM, Tark DS, Song JY, Hyun BH. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 15.Luo R, Fang L, Jin H, Jiang Y, Wang D, Chen H, Xiao S. Antiviral Res. 2011;91:99–101. doi: 10.1016/j.antiviral.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Jiang D, Qing M, Weidner JM, Qu X, Guo H, Chang J, Gu B, Shi PY, Block TM, Guo JT. Antiviral Res. 2009;83:53–60. doi: 10.1016/j.antiviral.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. J. Biol. Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 18.Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. J. Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 19.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. Proc. Natl. Acad. Sci. USA. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadler AJ, Williams BR. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sang Y, Rowland RR, Blecha F. J. Interferon Cytokine Res. 2010;30:1–7. doi: 10.1089/jir.2010.0016. [DOI] [PubMed] [Google Scholar]
- 22.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, Mcknight G, Clegg C, Foster D, Klucher KM. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 23.Song D, Park B. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Hayes J, Byrum B, Zhang Y. Virus Genes. 2016;52:578–581. doi: 10.1007/s11262-016-1334-x. [DOI] [PubMed] [Google Scholar]
- 25.Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Yoo D. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]


