Abstract
Hepatitis E Virus (HEV) is the major causative agent of acute hepatitis in developing countries. Its genome has three open reading frames (ORFs)—called as ORF1, ORF2, and ORF3. ORF1 encodes nonstructural polyprotein having multiple domains, namely: Methyltransferase, Y domain, Protease, Macro domain, Helicase, and RNA-dependent RNA polymerase. In the present study, we show that HEV-macro domain specifically interacts with light chain subunit of human ferritin (FTL). In cultured hepatoma cells, HEV-macro domain reduces secretion of ferritin without causing any change in the expression levels of FTL. This inhibitory effect was further enhanced upon Brefeldin-A treatment. The levels of transferrin Receptor 1 or ferroportin, two important proteins in iron metabolism, remained unchanged in HEV-macro domain expressing cells. Similarly, there were no alterations in the levels of cellular labile iron pool and reactive oxygen species, indicating that HEV-macro domain does not influence cellular iron homeostasis/metabolism. As ferritin is an acute-phase protein, secreted in higher level in infected persons and HEV-macro domain has the property of reducing synthesis of inflammatory cytokines, we propose that by directly binding to FTL, macro domain prevents ferritin from entering into circulation and helps in further attenuation of the host immune response.
Electronic supplementary material
The online version of this article (doi:10.1007/s11010-016-2715-0) contains supplementary material, which is available to authorized users.
Keywords: Open reading frame 1, Nonstructural protein, Macrodomain, Ferritin
Introduction
Hepatitis E Virus (HEV) is the major causative agent of acute inflammation of liver in developing countries. It is transmitted mainly by feco-oral route and occasionally by zoonotic infection [1]. HEV-infecting human, belongs to the family Hepeviridae, genus Orthohepevirus, and species Orthohepevirus A [2]. HEV virion is non-enveloped and spherical, exhibiting icosahedral symmetry. Its genome is a positive-sense RNA of ∼7.2 kb with a 5′-methylguanosine cap and a 3′ poly (A) stretch. It contains 5′ and 3′ Non-Coding Regions (NCRs) and three open reading frames (ORFs)—called as ORF1, ORF2, and ORF3, which encode nonstructural polyprotein, capsid protein, and a small phosphoprotein, respectively [3].
ORF1-encoded nonstructural polyprotein (pORF1) is approximately 1693 amino acid long and has multiple domains, namely: Methyltransferase, Y domain, Protease, X-domain (Macro domain), Helicase, and RNA-dependent RNA polymerase (RdRp) [4]. Functional characterization of individual domains has been done by cloning, partial purification, and in vitro biochemical assays. Amino acid (aa) region 1–979 of the pORF1 has been shown to be responsible for capping of HEV genomic RNA [5]. Protease domain, aa 433–592, deubiquitinates both RIG-I and TBK-1 proteins, and ubiquitination is known to be a key step in the activation of these proteins for poly (I:C)-induced type I interferon (IFN) synthesis [6]. Macro domain, aa 775–960, binds to poly (A) and poly ADP-ribose and catalyzes hydrolysis of ADP-ribose1″-phosphate [7]. Helicase domain, aa 960–1204, hydrolyzes nucleotide triphosphates and unwinds RNA duplexes with 5′ overhangs. It also possesses RNA 5′-triphosphatase activity [8, 9]. The RdRp domain, aa 1207–1693, can bind to 3′ end of HEV genomic RNA and has the ability to synthesize complementary RNA strand [10]. Although these proteins are functionally characterized, their role during virus replication and pathogenesis are still not well defined.
ADP-ribosylation is an important post-translational modification of proteins involved in many cellular processes such as DNA repair, genomic stability, transcriptional activation, and repression, to maintain telomere length, microtubule formation, centromere function, regulation of proteasomal protein degradation, regulation of endosomal vesicle trafficking, apoptosis etc. [11]. Macro domains are evolutionarily conserved throughout all eukaryotic organisms, bacteria and archaea [12]. Macro domains can bind to various ADP-ribose forms such as mono ADP-ribose, poly (ADP-ribose), poly(A), poly(G), O-acetyl-ADP-ribose [13], and can also catalyze the removal of proximal ADP-ribose residue [14, 15].
Among viruses, apart from HEV, macro domains are present in the members of families Coronaviridae and Togaviridae, as a part of nonstructural protein 3 (nsP3) [16]. These can bind to poly (A) and poly ADP-ribose groups and have the ability to hydrolyze ADP-ribose1″-phosphate [7]. Macro domains may vary in their binding affinities and forms of ADP-ribose bound [17]. In some coronaviruses, enzymatic activity of macro domains is dispensable during virus replication in cell culture [18–20], however, important during infections, as they play important role in restricting host innate immune response and pathogenesis [21–23]. While, in Sindbis virus, nsP3 macro domain is necessary for both replication as well as pathogenesis [24].
In HEV, macro domain is essential for genome replication only at the post-translational level [25] and not during the transcription phase [26]. Recently, Nan et al. [6] have shown the ability of HEV-macro domain to inhibit phosphorylation (activation) of interferon regulatory factor 3 (IRF-3), suggesting possible role in combating host antiviral response.
We have recently shown that protease, macro, and helicase domains of HEV pORF1 directly interact with light chain subunit of ferritin (FTL) using yeast two-hybrid system [27]. In the present study, we have analyzed the significance of FTL-macro domain interaction and its possible role during HEV pathogenesis.
Materials and methods
Yeast Two-Hybrid (Y2H) analysis
Materials used in Y2H experiment were purchased from Clontech Laboratories, Inc., Mountain View, CA, USA. They included culture media, yeast strains AH109 and Y187 and cloning vectors pGBKT7 and pGADT7 (as components of Matchmaker Two-Hybrid System 3, Cat. No. 630303) and X-α-Gal (Cat. No. 630407). Protocols for the yeast cell transformation, mating, and liquid β-galactosidase assay were carried out as per manufacturer’s instructions. Yeast strain AH109 transformed with pGBKT7-macro was used as “Bait” has been described previously [27]. PCR product encoding human Ferritin Light Chain (FTL) gene, generated from our previous study was cloned into pGADT7 vector and yeast cells (strain Y187) were transformed with this plasmid and used as “Prey”. These two yeast strains (with bait and prey) were subjected to hybridization and the yeast cells were plated onto quadruple dropout (QDO/Ade−, His−, Leu−, Trp−)/X-α-Gal selection plates and incubated at 30 °C for ~7 days. Colonies expressing reporter genes were selected and subjected to liquid β-galactosidase assay. Interaction between pGBKT7-lam and pGADT7-T antigen was used as a negative control while interaction between pGBKT7-p53 and pGADT7-T antigen served as a positive control.
Plasmids, antibodies and cell culture
Mammalian expression vector pcDNA3.1/myc-His(-) A (Cat No. V855-20) was from Thermo Fisher Scientific India, Pune, India. Green fluorescence protein expressing pAcGFP1-N1 (Cat. No. 632469) and Red fluorescence protein expressing pDsRedExpress-N1 (Cat No. 632429) plasmids were from Clontech Laboratories. HEV subgenomic replicon expressing Renilla luciferase (Rluc) gene (pSK-E2-Luc) was kindly provided by Dr. X.J. Meng (Virginia Tech, Blacksburg, VA, USA). Antibodies against c-Myc (Cat. No. M4439), α-tubulin (Cat. No. T6199) and HRP-conjugated secondary antibodies were from Sigma-Aldrich India, Bangalore, India. Pyridoxal Isonicotinoyl Hydrazone (PIH) (Cat. No. sc-204192), antibodies against GFP (Cat. No. sc-5385) and human Ferritin Light Chain protein (Cat. No. sc-74513) were from Santa Cruz Biotechnology Inc., Dallas, TX, USA. Anti-Ferroportin antibody (Cat. No. MTP11-A) was from Alpha Diagnostics, San Antonio, TX, USA and anti-Transferrin receptor 1 antibody (Cat. No. 136890) was from Thermo Fisher Scientific. Ferric Ammonium Citrate (FAC) (Cat. No. 0215804080) was from M.P. Biomedicals India, Mumbai, India.
Human hepatoma cells (Huh-7: S10-3) permissive for HEV replication were kindly provided by Dr. S. Emerson (NIH, Bethesda, MD, USA). Cells were grown in Dulbecco’s Modified Eagle’s medium supplemented with 10 % (vol/vol) heat-inactivated fetal calf serum, 100 U/ml penicillin, and 100 mg/ml streptomycin (Thermo Fisher Scientific). Cells were grown at 37 °C with 5 % CO2 in a humidified incubator. Transfections were performed using either Lipofectamine 2000 (Cat. No. 11668-019) or DMRIE-C (Cat. No. 10459014) (Thermo Fisher Scientific) transfection reagents as per the manufacturer’s instructions.
Co-affinity purification and co-localization
To verify in vivo physical interaction between HEV-macro domain and FTL protein, S10-3 (3 × 106 cells/well) were co-transfected with the expression constructs pcDNA-FTL and pAcGFP-macro. After 72 h, cells were harvested in RIPA buffer (50 mM Tris–Cl pH 8.0, 150 mM NaCl, 1 % IGEPAL CA-630, 0.5 % Sodium Deoxycholate, 0.1 % SDS) supplemented with protease inhibitor cocktail, complete mini EDTA-free (Product No. 11836170001, Roche India, Mumbai, India). Lysates were incubated with ProBond Nickel-Chelating Resin (Cat. No. R801-01, Thermo Fisher Scientific) for 2 h at 4 °C and processed further for the pull-down assay.
For co-localization, 5 × 105 cells were co-transfected with the expression constructs RFP-FTL and GFP-macro. After 72 h, cells were visualized under red as well as green filter using FLoid Cell Imaging Station (Thermo Fisher Scientific). Images were further processed by NIH Image software (ImageJ v1.50b; NIH, USA).
Quantitative PCR
To study the effect of HEV-macro domain on expression levels of different proteins involved in iron metabolism at transcriptional level, SYBR green-based quantitative reverse transcription PCR was carried out for selective genes (for primer sequences, see supplementary material). For that, 5 × 105 cells were transfected either with empty vector (pcDNA) (as a control) or with macro domain construct (pcDNA-macro) and harvested after 72 h. Total cellular RNA was extracted using Ribopure RNA extraction kit (Part No. AM1924, Thermo Fisher Scientific) and cDNA was prepared using High Capacity cDNA Reverse Transcription kit (Cat. No. 4368813, Thermo Fisher Scientific). Fifty nanogram of the cDNA (RNA equivalent) was used as a template and qPCR was performed using Kapa SYBR Fast qPCR Kit (Code KK4601, Kapa Biosystems, Inc., Wilmington, MA, USA). GAPDH was used as an endogenous control to normalize the input RNA.
Immunoblotting
Cellular proteins were extracted using RIPA buffer and protein content was quantitated by modified Lowry’s method. About 20 µg of protein was subjected to SDS-PAGE and transferred onto Nitrocellulose Membrane (Code No. RPN3032D, GE Healthcare India, Pune, India). After blocking overnight with 5 % non-fat dried milk powder in 1× PBS at 4 °C, the membranes were processed through sequential incubations with primary antibody followed by anti-primary IgG peroxidase-conjugated antibody. Immunoreactive proteins were visualized using Western Blotting detection Reagent (Code No. RPN2232, GE Healthcare). After stripping the primary antibodies with Restore Western Blot Stripping Buffer (Cat. No. 21059, Thermo Fisher Scientific), membranes were reprobed for α-tubulin, which served as a loading control for the normalization of densitometric values of the immunoblots. Signal intensities of blots were quantitated using ImageJ v1.50b software.
Ferritin ELISA
Measurement of ferritin protein in cell culture supernatant was carried out using Human Ferritin ELISA Kit (Cat. No. RAB0197, Sigma-Aldrich). Cells (5 × 105) were transfected either with empty vector or pcDNA-macro and cell culture supernatants were collected after 72 h. In the second set of transfected cells, Brefeldin-A (Cat No. B7651, Sigma-Aldrich) was added to the medium at 1 µg/ml concentration, 3 h prior to collection of supernatant.
Luciferase assay
Quantitation of HEV replication was done as described previously [28, 29]. Briefly, capped RNA transcripts of linearized plasmid pSK-E2-Luc were generated by mMessage mMachine T7 ultra kit (Cat. No. AMB1345-5, Thermo Fisher Scientific). Cells (5 × 105) were either left untransfected or co-transfected with HEV replicon and firefly luciferase-expressing plasmid (pGL4.10) by DMRIE-C transfection reagent. Cells were harvested at different time intervals and stored at −80 °C. Luciferase activity was measured using Dual Luciferase Reporter Assay System (Cat No. E1960, Promega Corporation, Madison, WI, USA). Luminiscence was measured on VictorX3 2030 Multilabel Reader (PerkinElmer).
Statistical analysis
Experiments were performed at least in triplicates. Data were analyzed by a two-tailed, unpaired Student’s t-test. Differences were considered significant at a p-value of ≤0.05.
Results
HEV-macro domain interacts with Ferritin Light Chain (FTL)
To confirm observations of HEV-macro domain–human FTL binary interactions from our previous Y2H experiments that were carried out using human liver cell cDNA library [27], we repeated Y2H experiment using individual clones. For that, yeast strains Y187 and AH109 were transformed with pGADT7-FTL (AD) and pGBKT7-HEV-macro (BD) encoding plasmids, respectively and hybridized (Fig. 1a). It was seen that the α-galactosidase reporter activity was specifically turned on only when HEV-macro domain and FTL co-existed in the hybrid cells, that also allowed growth of the hybrids on quadruple dropout medium (QDO/Ade−, His−, Leu−, Trp−). A quantitative estimation of relative strength of the interaction between HEV-macro domain and FTL was carried out using liquid β-galactosidase assay and it was observed that β-galactosidase levels of the HEV-macro–FTL hybrid cells were nearly 25 fold higher as compared to negative controls (Fig. 1b), indicating specificity of the interaction.
Fig. 1.
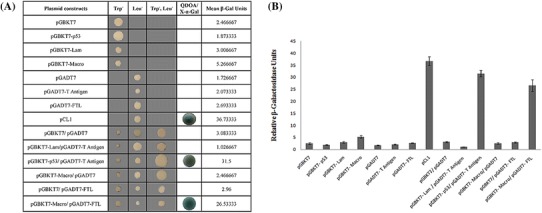
Yeast two-hybrid analysis of Macro–FTL interaction. a Yeast strain AH109 and Y187 were transformed with empty BD-(pGBKT7) and AD-(pGADT7) vectors or fused BD- and AD-expression constructs. Both strains were subjected to hybridization. BD-p53 and AD-T antigen served as positive control while BD-lamin and AD-T antigen were negative control. pCL1 is a plasmid which codes for full-length Gal-4p. Yeast cells were plated on synthetic dropout medium lacking Leucine and Tryptophan (Leu−, Trp−) as hybridization controls and to medium lacking Histidine (His−) and Adenine (Ade−) to select for yeast two-hybrid interactions at high (Leu−, Trp−, His−, Ade−) stringency with X-α-Gal as a substrate for reporter gene. Yeast plates were incubated at 30 °C for ~7 days. b Bar graph represents three independent experiments (mean ± SD)
To further confirm this interaction, human hepatoma cells (Huh-7: S10-3) were used as a model system as HEV replicates in hepatocytes. These cells were co-transfected with pcDNA-FTL (with c-Myc and His tags) and pAcGFP-macro clone plasmids and harvested after 72 h. Cell lysates were subjected to pull down using Ni2+ affinity resin and eluted proteins were probed with either anti-c-Myc or anti-GFP antibodies. FTL interacting HEV-macro was pulled down along with FTL (Fig. 2a). Since this pull-down experiment included both plasmid expressing proteins, to rule out the possibility of false interaction due to plasmid-based over expression, experiment was repeated by transfecting cells with pcDNA-macro construct alone. Macro domain also showed interaction with FTL expressed at the basal levels in these hepatoma cells (Fig. 2b). Overall, both experiments confirmed that HEV-macro domain specifically interacted with FTL.
Fig. 2.
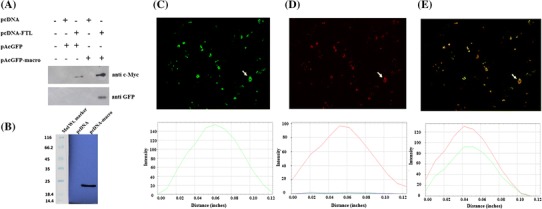
FTL protein interacts with HEV-macro domain in human hepatoma cells. a Huh-7 cells were co-transfected with empty vector plasmids or clones. After 72 h of transfection, cells were harvested and co-affinity purification was carried out using nickel affinity resin. Protein samples were subjected to SDS-10 % PAGE and immunoblotted with either anti-c-Myc or anti-GFP antibody. b HEV-macro domain co-precipitates with endogenous FTL. Cells were transfected with either pcDNA or pcDNA-macro and after 72 h of transfection, cells were harvested and co-affinity purification was carried out using nickel affinity resin. Protein samples were subjected to SDS-15 % PAGE and immunoblotted with anti-FTL antibody. Co-localization of HEV-macro domain and FTL protein. Huh-7 cells were co-transfected with GFP-macro and RFP-FTL clone plasmids. After 72 h of transfection, cells were visualized under green (c) as well as red filter (d) by FLoid Cell Imaging Station. Subsequently, both images were merged in (e). Images of single focal planes were processed using NIH Image software (ImageJ v1.50b; National Institutes of Health, USA). Profiles were calculated using the RGB profiler tool. (Color figure online)
After confirming the specificity of the protein–protein interaction, we decided to determine the localization of macro domain and FTL proteins in cells. For that, cells were co-transfected with pAcGFP-macro and pDsRedExpress–FTL plasmid constructs with Green and Red fluorescence tags, respectively. Expression of fusion proteins was confirmed by immunoblots (see Fig. 8 in the supplementary material) as well as observation using green and red filters in the FLoid cell imaging station (Fig. 2c, d). Image overlay analysis revealed that the two proteins are co-localized in cells, as seen by the yellow signals in the merged fields (Fig. 2e). Co-localization was also evident from the overlap of their respective profile plots (Fig. 2e). Taken together, results of Y2H, co-affinity purification, and co-localization experiments confirmed that HEV-macro domain interacts with human FTL protein.
HEV-macro domain marginally alters the expression of proteins of iron metabolism
Cellular iron homeostasis necessitates tight control of iron uptake, storage, and export and management of intracellular iron distribution. Under physiological conditions, extracellular iron in the circulation is bound to Transferrin (Tf), which is internalized by receptor-mediated endocytosis. After binding of Tf-iron complex to transferrin receptor 1 (TfR1), iron is released from Tf in cells. It is then reduced to ferrous ion (Fe2+) form and transported across endosomal membranes by a divalent metal transporter protein (DMT1), also known as Ferroportin (Fpn) and becomes part of the Labile Cell Iron (LCI) pool. Free iron can be utilized for metabolism, stored in oxidized form (Fe3+) in ferritin, or released back to extracellular space. Iron is potentially toxic as LCI catalyzes formation of reactive oxygen species (ROS) [30]. As liver stores substantial amount of iron in ferritin, interaction of HEV-macro domain with FTL compelled us to further investigate influence of HEV-macro domain on iron metabolism.
We analyzed levels of FTL, TfR1, and Fpn at the transcription and protein levels in cells expressing HEV-macro domain. HEV-macro domain did not exert any significant alterations in the transcription of those genes, as seen from the comparable levels of their transcripts in the cells transfected with pcDNA and pcDNA-macro (Fig. 3).
Fig. 3.
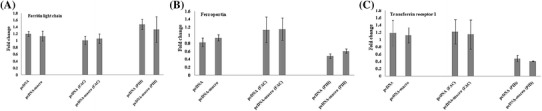
Hepatitis E virus macro domain marginally alters the transcription of iron metabolism genes. Huh-7 cells transfected by either empty vector or pcDNA-macro clone plasmid. After 72 h of transfection, cells were harvested and subjected to SYBR green-based quantitative reverse transcription PCR assays for a Ferritin light chain (FTL) b Ferroportin (Fpn) and c Transferrin receptor 1 (Tfr1) genes. Cells were either treated for 24 h with 100 µM Ferric Ammonium Citrate (FAC) or 100 µM Pyridoxal Isonicotinoyl Hydrazone (PIH) or left untreated. After normalization with GAPDH values, fold change in mRNA levels in transfected cells were expressed relative to control cells. The graphs represented three independent experiments (mean ± SD)
To understand the effect of macro domain in iron-depleted and iron-supplemented cells, we treated the transfected cells with either Ferric Ammonium Citrate (FAC) or Pyridoxal Isonicotinoyl Hydrazone (PIH), 24 h prior to harvesting. FAC is the known donor of Fe3+ ions and PIH is the chelator of labile iron pool in cultured hepatoma cells [31, 32]. Increase in Fe3+ conc. did not alter transcript levels in cells, either in the presence or absence of HEV-macro domain. While, depletion of free cellular iron in cells resulted in the lowering of Fpn and TfR1 transcript levels, HEV-macro domain itself did not have any influence on their expression levels (Fig. 3).
When analyzed at the protein levels by immunoblots, Fpn and FTL levels were found to be significantly reduced in iron-depleted cells, albeit, irrespective of the presence of HEV-macro domain. Further, though there were some changes in the expression levels of FTL in macro domain expressing cells, they were not statistically significant. The Fe3+ supplemented cells showed significant decrease (p = 0.0211) in TfR1 levels in presence of macro domain (Fig. 4) .
Fig. 4.
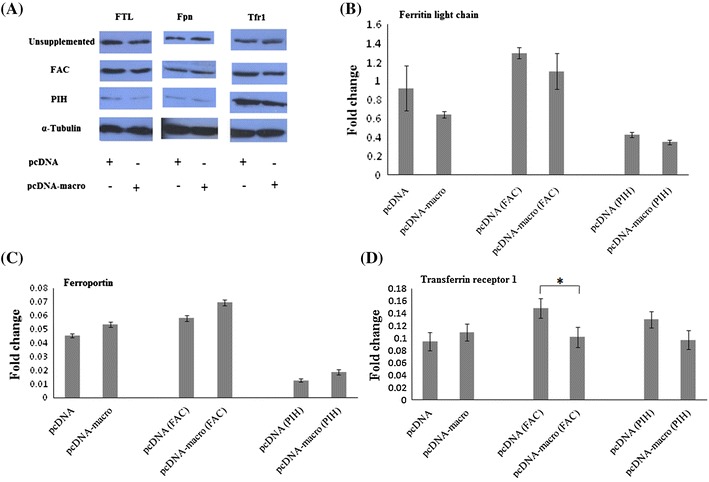
Hepatitis E virus macro domain marginally alters the expression of iron metabolism proteins. Huh-7 cells transfected by either empty vector or pcDNA-macro clone plasmid. After 72 h of transfection, cells were harvested and subjected to a immunoblot for Ferritin light chain (FTL), Ferroportin (Fpn), and Transferrin receptor 1 (Tfr1) proteins. Cells were either treated for 24 h with 100 µM Ferric Ammonium Citrate (FAC) or 100 µM Pyridoxal Isonicotinoyl Hydrazone (PIH) or left untreated. b–d shows fold change in signal intensities for FTL, Fpn, and Tfr1, respectively after normalization with α-tubulin. The graphs represented three independent experiments (mean ± SD, *p ≤ 0.05)
Measurement of Ferritin secretion
Cytosolic ferritin molecules are glycosylated and secreted by the classical secretory pathway. Ferritin molecules traverse endoplasmic reticulum (ER), pass through Golgi apparatus en route and get secreted [33]. We aimed to know whether macro domain has any effect on its secretion. Level of secreted ferritin in cell culture supernatant was determined by ELISA. HEV-macro domain showed significant reduction in the secretion of ferritin in normal, iron-supplemented as well as iron-depleted cells (Fig. 5). These results indicate that irrespective of the labile iron levels in cells, HEV-macro domain has inhibitory effect on ferritin secretion. This effect was further enhanced when cells were treated with Brefeldin-A, suggesting a different mechanism of inhibition by macro domain than Brefeldin-A-mediated inhibition, which occurs due to blockade of classical secretory pathway.
Fig. 5.
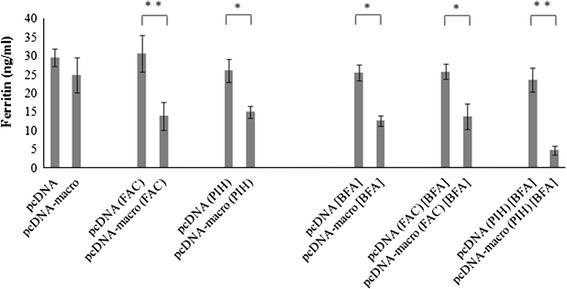
Measurement of ferritin secretion in the presence of HEV-macro domain. Huh-7 cells transfected by either empty vector or pcDNA-macro clone plasmid. Quantitation of ferritin was done by Enzyme Linked Immunosorbent Assay. Cells were either kept untreated or treated with 100 µM Ferric Ammonium Citrate (FAC) or 100 µM Pyridoxal Isonicotinoyl Hydrazone (PIH) for 24 h. In some experiments, cells were treated with Brefeldin-A (1 µg/ml) for 3 h. Values correspond to triplicate samples (mean ± SD, *p ≤ 0.05; **p ≤ 0.005)
To see whether inhibitory effect of macro domain on ferritin secretion is also seen during HEV replication, we used HEV subgenomic replicon expressing Renilla luciferase reporter gene (pSK-E2-Luc). Huh-7 (S10-3) cells were transfected with in vitro synthesized RNA transcripts and virus replication was monitored for up to 6 days by performing dual luciferase assay. Ferritin secretion was monitored by measuring ferritin in the culture supernatants. Renilla luciferase activity was increased consistently from 2 days onwards for up to 6 days, indicating successful virus replication. While, levels of secreted ferritin remained similar in both, mock transfected and HEV replicating cells (Fig. 6).
Fig. 6.
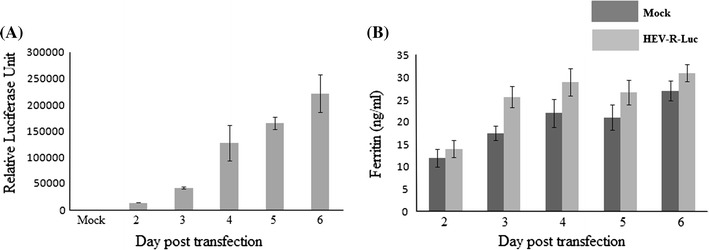
Measurement of ferritin secretion in the presence of subgenomic replicon of HEV. a Replication kinetics of HEV. Huh-7 cells were either left untransfected (mock) or co-transfected with capped RNA transcripts of pSK-E2-Luc and pGL4.10. At different days post transfection, cells were harvested and luciferase activity was measured. b Measurement of the secretion of intacellular ferritin in the presence of HEV replicon. Ferritin ELISA of the cell culture supernatant was performed at different time intervals. Values correspond to triplicate samples (mean ± SD)
Discussion
Hepatitis E virus is the main causative agent of acute viral hepatitis in developing countries. It is also an emerging zoonotic pathogen in several industrialized countries. For a long time, HEV remained the understudied virus due to the lack of efficient culture system and the unavailability of specific antibodies against viral proteins. Recently, using Y2H screening of human liver cDNA library, we identified 44 cellular proteins interacting with HEV-macro domain [27]. Among total interacting cDNA clones, human FTL clones were abundant (30 % of the interacting cDNA clones). As we know that, Y2H screening has a tendency to show false-positive protein–protein interactions, we confirmed HEV-macro domain–FTL binary interaction by repeating the Y2H experiment. This interaction was also confirmed in human hepatoma (Huh-7) cells by performing co-affinity purification and co-localization of these proteins. Hemopexin, another iron-binding protein of liver has been previously reported to interact with HEV-ORF3 protein [34].
Ferritin is a ubiquitous protein that stores iron in cells and releases it when cell needs it. By sequestering free iron in cells, ferritin serves dual function of iron detoxification and storage. It is composed of 24 subunits of light (FTL) and heavy (FTH) chain polypeptides with different proportion in different tissues [35]. Liver ferritin is mainly composed of light chain subunits of 175 amino acids [36, 37]. Viral macro domain proteins are known to bind to ADPR/poly ADPR moieties. As none of the amino acid residues in FTL are known to be ADP ribosylated, interaction between FTL and HEV-macro domain was intriguing. It indicated possible significance of this interaction in cellular iron metabolism.
Iron metabolism is controlled at different levels [38–40] and involves more than 100 genes with interlinked pathways [41]. In the current study, it was observed that though macro domain directly interacted with FTL, it did not alter the expression of key proteins of iron metabolism such as FTL, TfR1, and Fpn. Macro domain also did not affect cellular levels of labile iron or reactive oxygen species (data not shown) suggesting other role of macro domain–FTL interaction. However, a quantitative proteomics study of liver tissues of HEV-infected swine has shown altered expressions of proteins related to iron metabolism [42].
Iron Regulatory Proteins (IRPs) regulate the translation of FTL, TfR1, and Fpn proteins by binding to the Iron Responsive Elements (IREs) present in their cognate mRNAs [43]. Sequence analysis of HEV genome confirmed that there are no IREs in the viral genome and since clonal expression of HEV-macro domain did not influence expression levels of iron metabolism proteins, we did not carry out functional analysis of IRPs.
Iron plays an important role during viral infection. This element is required by both host and virus for their survival [44]. Alteration in expression of iron regulatory proteins during viral infections are reported [45–48]. Ferritin has been reported to play an important role in the pathogenesis of certain viruses. It protects shrimp Litopenaeus vannamei from white spot syndrome virus (WSSV) infection by inhibiting virus replication, presumably by reducing iron availability [49]. Protein kinase 1 of WSSV interacts with shrimp ferritin and prevents it from iron loading, and thus stabilizes the cellular labile iron pool. This may be an assault mechanism of WSSV to counteract the host cell’s iron-withholding defense mechanism [50]. Ferritin synthesis has been reported to be increased in mengovirus-infected cells to combat with the increased levels of free iron after virus infection [51]. Leader protein of mengovirus suppresses iron/ferritin-mediated NF-κB activation and subsequent suppression of alpha/beta interferon production in the infected cells [52].
HEV-macro domain inhibited the secretion of ferritin from human hepatoma cells. At the same time it did not significantly alter cellular levels of FTL. These observations remained same even in the altered cellular labile iron pools (iron supplementation and depletion). Secretion of ferritin takes place by the classical pathway, routed through the ER and golgi apparatus [33]. Brefeldin-A, an inhibitor of Golgi function, significantly reduced ferritin secretion from treated cells. HEV-macro domain and Brefeldin-A showed an additive inhibitory effect on ferritin secretion, suggesting that macro domain has a different mechanism for reducing the ferritin secretion.
Ferritin is an acute-phase protein and its elevated secretion is reported in the sera of acute viral hepatitis patients [53, 54]. Cytokines can alter synthesis of ferritin at transcription and/or posttranscription level [55, 56]. In rat hepatoma cells, interleukin-1 beta (IL-1β) or tumor necrosis factor-alpha (TNF-α) have been shown to stimulate ferritin secretion in the absence of any change in the intracellular ferritin levels [57]. In HepG2 (human hepatoma) cells, IL-1β and TNF-α stimulate synthesis of ferritin, particularly the FTL [58, 59]. These reports show the importance of cytokines in altering iron metabolism, though with different mechanisms. Ruddell et al. [60] have suggested that ferritin acts as a cytokine regulating pro-inflammatory function via NF-κB-regulated signaling in hepatic stellate cells.
HEV infection induces synthesis of pro-inflammatory chemokines, cytokines, and type 1 interferons in A549 cells (lung epithelial cell line) [61, 62]. TNF-α and IL-1β exposure can modulate iron uptake and ferritin synthesis in these cells [63]. HEV-macro domain has been shown to play an important role in downregulating type I IFN synthesis by inhibiting IRF-3 phosphorylation [6]. We suggest that, by direct binding to FTL, macro domain sequesters intracellular ferritin to prevent its secretion and attempts to restrain cellular innate immune response (Fig. 7).
Fig. 7.
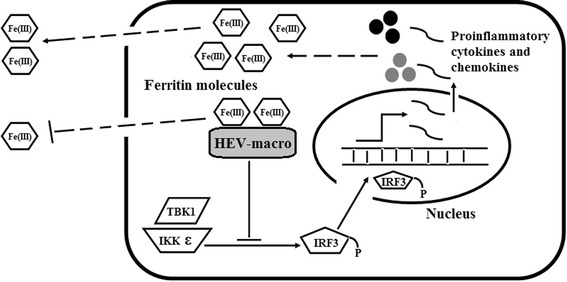
Schematic representation of how HEV-macro domain inhibits the secretion of intracellular ferritin
Intriguingly, helicase and protease domains of pORF1 were also found to interact with FTL and probably further contribute in reducing the ferritin secretion. To check this, we measured the secretion of ferritin by transfecting cells with HEV replicon, which expresses all the ORF1 domains. Irrespective of the increase in HEV replication (seen with Renilla luciferase activity), level of ferritin secretion remained constant for up to 6 days, suggesting important role of ORF1 domains in keeping check on ferritin secretion. It is necessary to analyze the significance of reduced ferritin secretion by HEV-macro domain. This would help in understanding importance of yet poorly characterized macro domain, apart from its canonical function of ADP-ribose binding, in HEV pathogenesis.
Conclusion
In the present study, by clonally expressing macro domain of HEV pORF1, we identified human light chain ferritin as an interacting partner of this protein. We also observed that macro domain alone was not sufficient to change expression levels of iron metabolism proteins such as TfR1, Fpn, and FTL. However, it has ability to inhibit secretion of ferritin. Cytokines seem to play an important role in ferritin turnover. Ferritin levels are known to increase in viral infections. Our study suggests that by directly interacting with FTL, HEV-macro domain inhibits its secretion and possibly suppresses innate immune response. Further, this interaction may also help in keeping the free iron level in cells, probably required for HEV genome replication. It may be a survival strategy of the virus. As indicated by the interactions of pORF1 domains with other proteins in iron metabolism, the viral intervention in the cellular iron metabolism could be complex and needs to be studied in an efficient cell culture model.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Primer sequences used in the present study and immunoblots indicating the expression of the clone plasmids, pcDNA-macro and pDsRedExpresN1-FTL. (DOCX 113 kb)
Acknowledgments
Funding to carry out this research was provided by the intramural grant of the National Institute of Virology, Pune (Grant ID: HEP 1104). N.K.O. received graduate research fellowship from the Council of Scientific and Industrial Research, Government of India (Grant ID: 09/698[0015]/2009-EMR-1). We thank Dr. S. Emerson (NIH, USA) and Dr. X.J. Meng (Virginia Tech, Blacksburg, USA) for providing Huh-7: S10-3 cell line and HEV-luciferase subgenomic replicon, respectively.
Compliance with ethical standards
Conflict of interest
Authors declare that there is no conflict of interest.
References
- 1.Perez-Gracia MT, Garcia M, Suay B, Mateos-Lindemann ML. Current knowledge on Hepatitis E. J Clin Transl Hepatol. 2015;3(2):117–126. doi: 10.14218/JCTH.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.http://www.ictvonline.org/
- 3.Emerson SU, Purcell RH. Hepatitis E virus. In: Knipe DM, Howley PM, editors. Fields virology. 6. Philadelphia: Lippincott Williams and wilkins Publishers; 2013. pp. 2242–2258. [Google Scholar]
- 4.Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magden J, et al. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol. 2001;75(14):6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol. 2014;88(20):11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egloff MP, Malet H, Putics A, et al. Structural and functional basis for ADP-ribose and poly (ADPribose) binding by viral macro domains. J Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpe YA, Lole KS. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol. 2010;84(18):9637–9641. doi: 10.1128/JVI.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpe YA, Lole KS. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol. 2010;84(7):3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal S, Gupta D, Panda SK. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp) Virology. 2001;282:87–101. doi: 10.1006/viro.2000.0819. [DOI] [PubMed] [Google Scholar]
- 11.Diefenbach J, Bürkle A. Introduction to poly(ADP-ribose) metabolism Cell. Mol Life Sci. 2005;62:721–730. doi: 10.1007/s00018-004-4503-3. [DOI] [PubMed] [Google Scholar]
- 12.Pehrson JR, Fuji RN. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutation Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenthal F, Feijs KL, Frugier E, et al. Macrodomain-containing proteins are new mono-ADPribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 16.Gorbalenya AE, Koonin EV, Lai MM. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288(1–2):201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst-Hess KR, Kuo L, Masters PS. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J Virol. 2015;89(11):6033–6047. doi: 10.1128/JVI.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putics A, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol. 2005;79(20):12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson KK, Cervantes-Barragan L, Ludewig B, Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89(3):1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuri T, Eriksson KK, Putics A, et al. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J Gen Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 24.Park E, Griffin DE. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology. 2009;388:305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvez MK. The hepatitis E virus ORF1 ‘X-domain’ residues form a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene. 2015;566(1):47–53. doi: 10.1016/j.gene.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvez MK. Molecular characterization of hepatitis E virus ORF1 gene supports a papain-like cysteine protease (PCP)-domain activity. Virus Res. 2013;178:553–556. doi: 10.1016/j.virusres.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ojha NK, Lole KS. Hepatitis E virus ORF1 encoded non structural protein-host protein interaction network. Virus Res. 2016;213:195–204. doi: 10.1016/j.virusres.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Cao D, Huang YW, Meng XJ. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol. 2010;84(24):13040–13044. doi: 10.1128/JVI.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devhare P, Sharma K, Mhaindarkar V, Arankalle V, Lole K. Analysis of helicase domain mutations in the hepatitis E virus derived from patients with fulminant hepatic failure: effects on enzymatic activities and virus replication. Virus Res. 2014;184:103–110. doi: 10.1016/j.virusres.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane DJ, Merlot AM, Huang ML, et al. Cellular iron uptake, trafficking and metabolism: key molecules and mechanisms and their roles in disease. Biochim Biophys Acta. 2015;5:1130–1144. doi: 10.1016/j.bbamcr.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Hirsh M, Konijn AM, Iancu TC. Acquisition, storage and release of iron by cultured human hepatoma cells. J Hepatol. 2002;36(1):30–38. doi: 10.1016/S0168-8278(01)00221-5. [DOI] [PubMed] [Google Scholar]
- 32.Ponka P, Richardson D, Baker E, Schulman HM, Edward JT. Effect of pyridoxal isonicotinoyl hydrazone and other hydrazones on iron release from macrophages, reticulocytes and hepatocytes. Biochim Biophys Acta. 1988;967(1):122–129. doi: 10.1016/0304-4165(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S, Hevi S, Chuck SL. Regulated secretion of glycosylated human ferritin from hepatocytes. Blood. 2004;103(6):2369–2376. doi: 10.1182/blood-2003-09-3050. [DOI] [PubMed] [Google Scholar]
- 34.Ratra R, Kar-Roy A, Lal SK. The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry. 2008;47(7):1957–1969. doi: 10.1021/bi7016552. [DOI] [PubMed] [Google Scholar]
- 35.Arosio P, Ingrassia R, Cavadini P. Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta. 2009;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Addison JM, Fitton JE, Lewis WG, May K, Harrison PM. The amino acid sequence of human liver apoferritin. FEBS Lett. 1983;164:139–144. doi: 10.1016/0014-5793(83)80037-4. [DOI] [PubMed] [Google Scholar]
- 37.http://www.uniprot.org/
- 38.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51(29):5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva B, Faustino P. An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim Biophys Acta. 2015;1852(7):1347–1359. doi: 10.1016/j.bbadis.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muckenthaler M, Richter A, Gunkel N, et al. Relationships and distinctions in iron-regulatory networks responding to interrelated signals. Blood. 2003;101(9):3690–3698. doi: 10.1182/blood-2002-07-2140. [DOI] [PubMed] [Google Scholar]
- 42.Lee G, Han D, Song J-Y, Kim J-H, Yoon S. Proteomic analysis of swine hepatitis E virus (sHEV)-infected livers reveals upregulation of apolipoprotein and downregulation of ferritin heavy chain. FEMS Immunol Med Microbiol. 2011;61:359–363. doi: 10.1111/j.1574-695X.2010.00770.x. [DOI] [PubMed] [Google Scholar]
- 43.Anderson CP, Shen M, Eisenstein RS, Leibold EA. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim Biophys Acta. 2012;1823(9):1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 45.Feng WR, Zhang M, Su YQ, Wang J, Wang YT, Mao Y. Identification and analysis of a Marsupenaeus japonicus ferritin that is regulated at the transcriptional level by WSSV infection. Gene. 2014;544(2):184–190. doi: 10.1016/j.gene.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 46.Fillebeen C, Muckenthaler M, Andriopoulos B, et al. Expression of the subgenomic hepatitis C virus replicon alters iron homeostasis in Huh7 cells. J Hepatol. 2007;47(1):12–22. doi: 10.1016/j.jhep.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 47.Maffettone C, De Martino L, Irace C, et al. Expression of iron-related proteins during infection by bovine herpes virus type-1. J Cell Biochem. 2008;104(1):213–223. doi: 10.1002/jcb.21618. [DOI] [PubMed] [Google Scholar]
- 48.Tsuji Y, Kwak E, Saika T, Torti SV, Torti FM. Preferential repression of the H subunit of ferritin by adenovirus E1A in NIH-3T3 mouse fibroblasts. J Biol Chem. 1993;268(10):7270–7275. [PubMed] [Google Scholar]
- 49.Ye T, Wu X, Wu W, Dai C, Yuan J. Ferritin protect shrimp Litopenaeus vannamei from WSSV infection by inhibiting virus replication. Fish Shellfish Immunol. 2015;42(1):138–143. doi: 10.1016/j.fsi.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 50.Lin SJ, Lee DY, Wang HC, et al. White spot syndrome virus protein kinase 1 defeats the host cell’s iron-withholding defense mechanism by interacting with host ferritin. J Virol. 2015;89(2):1083–1093. doi: 10.1128/JVI.02318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulvey MR, Kühn LC, Scraba DG. Induction of ferritin synthesis in cells infected with Mengo virus. J Biol Chem. 1996;271(16):9851–9857. doi: 10.1074/jbc.271.16.9851. [DOI] [PubMed] [Google Scholar]
- 52.Zoll J, Melchers WJ, Galama JM, van Kuppeveld FJ. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-kappa B. J Virol. 2002;76(19):9664–9672. doi: 10.1128/JVI.76.19.9664-9672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hengeveld P, Zuyderhoudt FM, Jöbsis AC, van Gool J. Some aspects of iron metabolism during acute viral hepatitis. Hepatogastroenterology. 1982;29(4):138–141. [PubMed] [Google Scholar]
- 54.Kotoh K, Ueda A, Tanaka M, et al. A high prevalence of extreme hyperferritinemia in acute hepatitis patients. Hepat Med. 2009;14(1):1–8. doi: 10.2147/hmer.s4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mascotti DP, Rup D, Thach RE. Regulation of iron metabolism: translational effects mediated by iron, heme, and cytokines. Annu Rev Nutr. 1995;15:239–261. doi: 10.1146/annurev.nu.15.070195.001323. [DOI] [PubMed] [Google Scholar]
- 56.Rogers J, Lacroix L, Durmowitz G, Kasschau K, Andriotakis J, Bridges KR. The role of cytokines in the regulation of ferritin expression. Adv Exp Med Biol. 1994;356:127–132. doi: 10.1007/978-1-4615-2554-7_14. [DOI] [PubMed] [Google Scholar]
- 57.Tran TN, Eubanks SK, Schaffer KJ, Zhou CY, Linder MC. Secretion of ferritin by rat hepatoma cells and its regulation by inflammatory cytokines and iron. Blood. 1997;90(12):4979–4986. [PubMed] [Google Scholar]
- 58.Hirayama M, Kohgo Y, Kondo H, Shintani N, Fujikawa K, Sasaki K, Kato J, Niitsu Y. Regulation of iron metabolism in HepG2 cells: a possible role for cytokines in the hepatic deposition of iron. Hepatology. 1993;18(4):874–880. doi: 10.1002/hep.1840180420. [DOI] [PubMed] [Google Scholar]
- 59.Rogers JT, Bridges KR, Durmowicz GP, Glass J, Auron PE, Munro HN. Translational control during the acute phase response. Ferritin synthesis in response to interleukin-1. J Biol Chem. 1990;265(24):14572–14578. [PubMed] [Google Scholar]
- 60.Ruddell RG, Hoang-Le D, Barwood JM, Rutherford PS, Piva TJ, Watters DJ, Santambrogio P, Arosio P, Ramm GA. Ferritin functions as a proinflammatory cytokine via iron-independent protein kinase C zeta/nuclear factor kappaB-regulated signaling in rat hepatic stellate cells. Hepatology. 2009;49(3):887–900. doi: 10.1002/hep.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devhare PB, Chatterjee SN, Arankalle VA, Lole KS. Analysis of antiviral response in human epithelial cells infected with hepatitis E virus. PLoS One. 2013;8(5):e63793. doi: 10.1371/journal.pone.0063793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong C, Zafrullah M, Mixson-Hayden T, Dai X, Liang J, Meng J, Kamili S. Suppression of interferon-α signaling by hepatitis E virus. Hepatology. 2012;55(5):1324–1332. doi: 10.1002/hep.25530. [DOI] [PubMed] [Google Scholar]
- 63.Smirnov IM, Bailey K, Flowers CH, Garrigues NW, Wesselius LJ. Effects of TNF-alpha and IL-1beta on iron metabolism by A549 cells and influence on cytotoxicity. Am J Physiol. 1999;277(2 Pt 1):L257–L263. doi: 10.1152/ajplung.1999.277.2.L257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used in the present study and immunoblots indicating the expression of the clone plasmids, pcDNA-macro and pDsRedExpresN1-FTL. (DOCX 113 kb)


