Abstract
The nucleocapsid (N) gene of the porcine epidemic diarrhea virus (PEDV) strain LJB/03 which was previously isolated in Heilongjiang province, China, was cloned, sequenced and compared with published sequences of other avian and mammalian coronavirus. The nucleotide sequence encoding the entire N gene open reading frame (ORF) of LJB/03 was 1326 bases long and encoded a protein of 441 amino acids with predicted Mr of 49 kDa. It consisted of 405 adenines (30.5%), 294 cytosines (22.1%), 329 guanines (24.8%) and 298 thymines (22.5%) residues. Sequence comparison with other PEDV strains selected from GenBank revealed that the LJB/03 N gene has a high sequence homology to those of other PEDV isolates, 97.4% with JS2004, 95.6% with chinju99, 96.6% with Br1/87, and 96.8% with CV777. The encoded protein shared 96.4% amino acid identities compared with CV777, 96.1% with Brl/87, 98% with JS2004, 96.90% with chinju99, respectively. The amino acid sequence contained seven potential protein kinase C phosphorylation sites, nine Casein kinase II phosphorylation sites, one Tyrosine kinase phosphorylation site, two cAMP- and cGMP-dependent protein kinase phosphorylation sites.
Keywords: Nucleocapsid (N) gene, Nucleotide sequence, PEDV
Introduction
Porcine epidemic diarrhea virus (PEDV) is classified as a member of the vCoronaviridae and causes acute enteritis in pigs [1], which was first reported in England in 1971 and has been reported in many countries such as Germany, Canada, Japanese, Korea, France, Belgium, Switzerland, etc. [2–5]. The disease was first reported in China in 1976 [6], caused serious economic losses due to the death of neonatal piglets and weight loss of the infected pigs. Clinical signs of PED include anorexia, vomiting, diarrhea, and dehydration. Morbidity and mortality in infected neonatal piglets less than 5 days old approach 100% because of severe diarrhea and dehydration. However, mortality in infected piglets older than 10 days is less than 10% [1].
The coronavirus genome consists of a positive-sense, single-stranded RNA molecule that is 20–30 kb in size [7]. Virions are enveloped, pleomorphic, and 80–220 nm in diameter, and they have club-shaped peplomers approximately 20 nm in length. Coronavirus possesses four major structural proteins including a phosphorylated nucleocapsid (N) protein and three envelope proteins, membrane protein (M), spike protein (S), and envelope protein (E); the first two envelope proteins are major envelope proteins, while the amount of E protein in virion is low [8–10], S glycoprotein makes up the large surface projections of the virion and the M and E proteins are essential for viral envelope formation and release [11, 12].
Studies indicate that the N proteins of coronaviruses are extensively phosphorylated, highly basic, and binds, to the viral genomic RNA forming a helical ribonucleoprotein (RNP) [13]. A variety of functional activities have been ascribed to the N proteins of previously known coronaviruses, including participation in transcription of the viral genome, the formation of viral core, and packaging viral RNA [14]. The N protein is highly immunogenic, more then, the cellular immune response against N protein of some animal coronaviruses can enhance the recovery from the virus infection [15, 16].
N protein can accumulates intra-cellularly even before it is packed in the mature virus [17] and is the most abundant virus derived-protein throughout the infection, probably because its template mRNA is the most abundant subgenomic RNA [18]. These features make it a suitable candidate for the accurate and early diagnosis and develop genetically engineered vaccines [19].
The aim of present study was to determine the complement nucleotide sequence of the PEDV N gene and get more information about PEDV isolates comes from different region. In this study, the RNA of PEDV was extracted directly from the feces samples of piglets naturally infected with PEDV LJB/03. The N gene has been cloned, sequenced and compared with other PEDV strains. These data are useful for further the study of molecular biology of PEDV strains that are prevalent in China.
Materials and methods
Virus strain
PEDV LJB/03 was collected from the feces of piglets suffering from severe diarrhea in HeiLongJiang, China.
Viral RNA extraction
The feces sample was operated following the methods of Fan Jinghui and Li Yijing [20]. The feces sample was diluted 1–10 in a disruption buffer (500 mM Tris–HCI [pH 8.3], 2% (w/v) PVP-40, 1% (w/v) PEG6000, 140 mM NaCl, 0.05% (v/v) Tween 20), vortexed, incubated at room temperature for 10 min, and centrifuged using a Beckman F3602 rotor at 2000 × g at 4°C for 5 min. The supernatant was removed and used for the extraction of the viral RNA using the Trizol reagent (Invitrogen USA) according to the manufacturer’s protocol and dissolved in diethyl procarbonate-treated distilled water.
Primers for RT-PCR
A pair of sense and antisense primer was designed and aligned based on nucleotide sequences of the N gene of CV777 and Brl/87 available in GenBank. The sense primer 5′-TTATGGCTTCTGTCAGCTTT-3′ and antisense primer 5′-ACATTGTTTAATTTCCTGTATC-3′ were used to amplify the n gene coding for the N protein of PEDV strain LJB/03.
Reverse transcriptase polymerase chain reaction
Synthesis of the first-strand cDNA for n gene was carried out by reverse transcription using promega reverse transcription reagent. The viral RNA (50 μl) was mixed with 2.5 μl of 10 pM of the antisense primer, incubated at 65°C for 5 min, and then placed on ice for 2 min. After that, 4 μl of 5× RT buffer, 4 μl of 2.5 mM dNTP mixture, 1 μl of RNase inhibitor (40 U/μl), 1 μl of reverse transcriptase (200 U/μl), 2.5 μl H2O was added and mixed gently. The reaction mixture was incubated for 50 min at 42°C, and was terminated by heating for 10 min at 65°C. RNase H (1 μl) was added to degrade RNA template for 20 min at 37°C prior to PCR amplification.
PCR was carried out in a 50 μl volume by mixing the cDNA above with 2.5 μl of each 10 pM sense and antisense, 1 mM each of dATP, dGTP, dTTP, dCTP, 5 μl of 10× PCR buffer (100 mM Tris–HCl, 1.5 mM MgC12, 50 mM KC1, pH 8.3), and 2.5 U Taq DNA polymerase (TaKaRa Biotechnology (Dalian) Co. Ltd.). Cycles were as follows: 94°C for 15 s, followed by 30 cycles of 94°C for 40 s, 49°C annealing for 30 s, 72°C extension for 1 min and a final extension of 72°C for 5 min.
The PCR product was analyzed by electrophoresis through an agarose gel (Fig. 1), and visualized by staining with ethidium bromide, the target cDNA band was extracted from the gel using the Qiagen® gel extraction kit according to the manufacturer’s instructions. The purified PCR products were cloned into the pGEM-T easy vector (Promega, Madison, USA) with T4 DNA ligase. The plasmids were transformed into E. coli DH5α using standard molecular technique. Plasmid DNA was extracted by alkaline-lysis from E. coli DH5α culture and verified by using restriction enzyme digestion, PCR and electrophoresis in 1% agarose (Fig. 2). Colonies with correct sizes was named pGEM-T-N and at least three independent plasmid clones were analyzed, confirmed and sequenced.
Fig. 1.
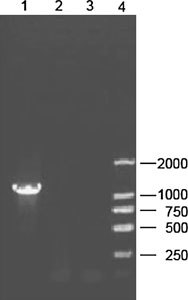
PCR products of PEDV N gene on 1% gel. Lane 1, N products (1326 bp); Lane 2 and 3, negative control; Lane 4, a molecular weight Marker DL 2000
Fig. 2.
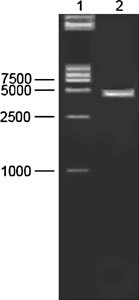
Restriction enzyme analysis of the recombinant plasmid pGEM-T-N. Lane 1, Marker DL 15000; Lane 2, digestion of the recombinant pGEM-T-N DNA (3400 bp) with EcoR I
Sequencing
The nucleotide sequence of the N gene of LJB/03 was, determined by TaKaRa Biotechnology (Dalian) Co. Ltd.
Sequence analysis
Amino acid sequences were aligned using the CLUSTAL W method, and phylogenetic trees were constructed using the neighbor-joining method. Analyses were done using the MegAlign application of the Lasergene software package. The identification of sequence motifs was done with the Psi-Blast program using the Swiss-Prot database through the MyHits web server (http://myhits.isb-sib.ch).
Results
By using RT-PCR method, we successfully amplified the nucleocapsid gene. The PCR products were approximately 1.3 kb in size and cloned into the pGEM-T easy vector.
The complete nucleotide sequence of nucleocapsid gene has been deposited in GenBank, accession number is DQ072726.
Sequence analysis indicate that the compete open reading frame (ORF) for the nucleocapsid gene of PEDV LJB/03 consists of 1326 bases and codes for a basic protein of 441 amino acid. It consisted of 405 adenines (30.5%), 294 cytosines (22.2%), 329 guanines (24.8%) and 298 thymines (22.5%) and a G+C content of 47.0%.
The result of motif blast indicated the LJB/03 N protein had seven potential protein kinase C phosphorylation sites, nine Casein kinase II phosphorylation sites, one Tyrosine kinase phosphorylation site, two cAMP- and cGMP-dependent protein kinase phosphorylation sites.
The gene had 43 nucleotide mismatches compared to CV777, a substantial portion (72%, 31/43) of the substitutions was transversions, about 60% of the substitutions were non-synonymous mutations.
Table 1 shows that the percent similarity of the N nucleotide sequences varied from 95.6% to 97.4% between LJB/03 and the other four strains of PEDV, and a high degree of identity (94.9–99.8%) was observed between the nucleotide sequences of PEDV strains. The alignment of the nucleotide sequences shows that no deletion or insertion event was detected, and there is a large region of absolute identity such as in the region from nucleotide 517 to nucleotide 614 (517–614 bases).
Table 1.
Comparision of the nucleotide and deduced amino acid of the N gene of PEDV LJB/03 and CV777, Br1/87, chinju99, JS2004
| Homology percent—amino acid | |||||
|---|---|---|---|---|---|
| LJB/03 | Brl/87 | Chinju99 | CV777 | JS2004 | |
| LJB03 | 96.1 | 95.9 | 96.4 | 98.0 | |
| Br1/87 | 96.6 | 96.6 | 99.8 | 95.9 | |
| chinju99 | 95.6 | 96.3 | 96.8 | 95.2 | |
| CV777 | 96.8 | 99.8 | 96.5 | 96.1 | |
| JS2004 | 97.4 | 95.6 | 94.9 | 95.8 | |
| Similarity percent—nucleotides | |||||
The entire nucleocapsid protein of PEDV LJB/03 aligned with the published sequences of CV777, Brl/87, chinju99 and JS2004. This alignment indicates that overall the sequences are, highly conserved with some regions showing no variation at all, and the 15 nucleotide acid substitutions in the 5′ region (1–249 bases) did not arouse amino acid changes, which may suggestion the N-terminal of the protein had more homologous than the C-terminal. Two-way comparisons among the nucleocapsid proteins of these five strains of PEDV indicate that the identities range from 95.9% to 98.0%, with CV777 and Brl/87 having the most identity, and LJB/03 and chinju99 the least.
Phylogenetic analysis
A phylogenetic tree was prepared to further examine relationships between PEDV and other coronaviruses based on a comparison of N protein amino acid sequences (Fig. 3). Phylogenetic analysis showed that PEDV was more closely related to group 1 (TGEV, HCV 229E and FIPV) than to members of group 2 coronaviruses (BCV, HCV OC43, and MHV) and group 3 (IBV and TCV) coronaviruses. In addition, phylogenetic analyses based on N protein sequences demonstrated that PEDV strains LJB/03 and JS2004 which comes from China and chinju99 comes from Korea are more closely related to each other than they are to those two isolates European CV777 and Brl/87.
Fig. 3.
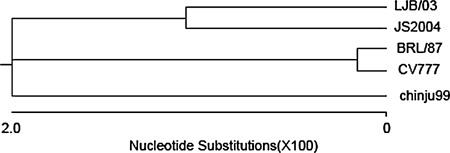
Phylogenetic relationship of PEDV LJB/03 and other PEDV strains based on a comparison of N protein sequences. Amino acid sequences were aligned using the CLUSTAL method, and phylogenetic. trees were constructed using the neighbor-joining method. Analyses were done using the MegAlign application of the Lasergene software GenBank accession numbers of sequences in the phylogenetic tree are: LJB/03 DQ072726; chinju99 AF237764; CV777 NC003436; Brl/87 Z24733 (Britain isolate); JS2004 AY653206 (China field isolate)
Discussion
In the present study, the N gene of LJB/03 was cloned and sequenced. The result sequence revealed the N gene has a ORF of 1326 nucleotides coding for a 441 amino acids protein. Sequence comparison with other PEDV strains selected from GenBank indicated that the N gene of PEDV was highly conserved even though comes from different geographic region, and the alignment result showed there is some region of absolute identity in the sequences.
Previous studies showed the chinju99 N protein had 7 potential T- or S-linked phosphorylation sites and seven potential Casein kinase II phosphorylation sites, the result in this study indicated the LJB/03 N protein had seven potential protein kinase C phosphorylation sites, nine Casein kinase II phosphorylation sites, one Tyrosine kinase phosphorylation site, two cAMP- and cGMP-dependent protein kinase phosphorylation sites.
The entire nucleocapsid protein of PEDV LJB/03 aligned with the published sequences of CV777, Brl/87, chinju99 and JS2004. This alignment of nucleocapsid protein sequences indicates that overall the sequences are highly conserved with some regions showing no variation at all. This can be the feasible information for the development of genetically engineered N protein for vaccine to prevent PEDV infections. Shuichi et al. developed a method of detection of PEDV Using Polymerase Chain Reaction based on part of nucleocapsid nucleotide, and then compare the Nucleocapsid nucleotide among Strains of the Virus, the result of restriction analysis the PCR products were that CV777 and all the Korean strain can be digested with Dra I, EcoR I, but the Korean strain was not digested with Pst I. We found the N gene of LJB/03 and JS2004 (another China isolate) have the same restriction patterns with the Korean strains [21].
Coronaviruses have been subdivided into three major antigenic groups based on antigenic differences identified by serological analyses and nucleotide sequence analyses [22, 23]. Group I members are the porcine transmissible gastroenteritis virus (TGEV) and epidemic diarrhea virus (PEDV), feline and canine coronavirus (FCoV and CCoV), and human coronavirus 229E (HCoV-229E). Group II includes porcine hemagglutinating encephalomyelitis virus (HEV), murine hepatitis virus (MHV), bovine, equine, and rat coronavirus (BCoV, ECoV, and RtCoV), and human coronavirus OC43 (HcoV-OC43). Group III is specific for avian species including turkey coronavirus (TCoV), pheasant coronavirus and avian infectious bronchitis virus (IBV). The coronavirus N protein has been shown to be highly variable in size as well as in amino acid composition between the viruses that comprise the three coronavirus antigenic groups but highly conserved within these groups. Group I viral genomes have the smallest nucleocapsid protein with 378–389 residues, group II genomes have the largest with 449–455 residues and Group III 409 residues. All 5 PEDV strains had 441 amino acid residues, and have a longer peptide than other Group I members, which illuminate PEDV, a particular case is an exception to the rule of the coronavirus N protein has been shown to be highly conserved within these groups.
In the study, we acquired the nucleotide sequence of the N gene PEDV LJB/03 and did the nucleotide sequence analysis to establish the phylogenetic relationships between several strains of PEDV. This work showed that the nucleotide sequence can form a base for further study on the epidemiological study of PEDV infections.
Acknowledgement
The financial support of this work was provided by grants from “project of the tenth-five” of Heilongjiang provincial scientific and technique committee, China.
References
- 1.M.B. Pensaert, in: Diease of Swine, eds. by B.E. Straw, S. D’Aallaire, W.L. Mengeling, D.I. Taylor (The Lowa University Press, Ames, IA, 1999), pp. 179–185
- 2.J. Oldham, Pig Farming (Oct Suppl), 1972, pp. 72–73
- 3.DeBouck P., Callebaut P., Pensaert M. Proc. Int. Congr. Pig. Vet. Soc. 1982;7:53. doi: 10.1016/0378-1135(82)90009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann M., Wyler R. Schweiz Arch Tierheilkd. 1987;129:437. [PubMed] [Google Scholar]
- 5.Mostl K., Horvath E., Burki F. Wien Tierarztl Monatsschr. 1990;77:10. [Google Scholar]
- 6.Qinghua C., Xiaoying N., Chengyu Y. J. Anim. Vet. Sci. 1992;22(3):22. [Google Scholar]
- 7.Lai M.M., Cavanagh D. Adv. Virus Res. 1997;48:1. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godet M., L’Haridon R., Vautherot J.F., Laude H. Virology. 1992;188(2):666. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith A.R., Boursnell M.E., Binns M.M., Brown T.D. J. Gen. Virol. 1990;71(Pt 1):3. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu X., Bi W., Weiss S.R., Leibowitz J.L. Virology. 1994;202:1018. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- 11.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. EMBO J. 1996;15(8):2020. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. Virology. 1996;218(1):52. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macneughton M.R., Davies H.A. J. Gen. Virol. 1978;39(3):545. doi: 10.1099/0022-1317-39-3-545. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Zhang X. Virology. 1999;265:96. doi: 10.1006/viro.1999.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glansbeek H.L., Haagmans B.L., Lintelo E.G., Egberink H.F., Duquesnem V., Aubert A., et al. J. Gen. Viro1. 2002;83:1. doi: 10.1099/0022-1317-83-1-1. [DOI] [PubMed] [Google Scholar]
- 16.Wesseling J.G., Godeke G.J., Schijns V.E., Prevec. L., Graham F.L., Horzinek M.C., et al. J. Gen. Virol. 1993;74:2061. doi: 10.1099/0022-1317-74-10-2061. [DOI] [PubMed] [Google Scholar]
- 17.Collisson E.W., Pei J., Dzielawa J., Seo S.H. Dev. Comput. Immunol. 2000;24:187. doi: 10.1016/S0145-305X(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 18.Hiscox J.A., Caavanagh D., Britton P. Virus Res. 1995;36:119. doi: 10.1016/0168-1702(94)00108-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H. K., Yeo S.-G. Virus Genes. 2003;26:207. doi: 10.1023/A:1023447732567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J., Li Y. Virus Genes. 2005;30:69. doi: 10.1007/s11262-004-4583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubota S., Sasaki O., Amimoto K., Okada N., Kitazima T., Yasuhara H. J. Vet Med. Sci. 1999;61(7):827. doi: 10.1292/jvms.61.827. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez J.M., Gomez P., Cavanagh D., Gorbalenya A.E., Enjuanes L. Arch. Virol. 2003;48:2207. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya A.E., Snijder E.J., Spaan W.J. J. Virol. 2004;78:7863. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


