Abstract
Bats may be natural reservoirs for a large variety of emerging viruses, including mammalian coronaviruses (CoV). The recent emergence of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) in humans, with evidence that these viruses may have their ancestry in bats, highlights the importance of virus surveillance in bat populations. Here, we report the identification and molecular characterization of a bat β-Coronavirus, detected during a viral survey carried out on different bat species in the island of Sardinia (Italy). Cutaneous, oral swabs, and faecal samples were collected from 46 bats, belonging to 15 different species, and tested for viral presence. Coronavirus RNA was detected in faecal samples from three different species: the greater horseshoe bat (Rhinolophus ferrumequinum), the brown long-eared bat (Plecotus auritus), and the European free-tailed bat (Tadarida teniotis). Phylogenetic analyses based on RNA-dependent RNA polymerase (RdRp) sequences assigned the detected CoV to clade 2b within betacoronaviruses, clustering with SARS-like bat CoVs previously reported. These findings point to the need for continued surveillance of bat CoV circulating in Sardinian bats, and extend the current knowledge on CoV ecology with novel sequences detected in bat species not previously described as β-Coronavirus hosts.
Keywords: Bats, Coronavirus, RNA-dependent RNA polymerase, Rhinolophus ferrumequinum, Sardinia
Introduction
Bats (order Chiroptera) are one of the most diverse and widely distributed group of mammals, comprising more than 1300 species across 21 families [1], and play a key role in terrestrial ecosystems [2]. In the last few decades, they have been associated with several emergent zoonotic agents and recognized as potential reservoir of zoonotic viruses belonging to different families, such as Hendra, Nipah, Ebola, Lyssa, and SARS Coronavirus [3]. Bats represent ideal hosts and reservoirs of viruses, due to their high diversity and ecological characteristics that potentially facilitate virus transmission, such as the roosting together behavior and their ability to fly over distance [4, 5]. Therefore, it is essential to identify and characterize the currently unrecognized viruses circulating in bat populations.
Coronaviruses (CoV, family Coronaviridae) are enveloped viruses with large single-stranded RNA genomes ranging between 27 and 32 kb [6, 7]. Based on genome-scale phylogenies, they are classified into four genera: Alphacoronavirus (α-CoV) and Betacoronavirus (β-CoV), pathogenic for mammals, Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV), primarily (but not exclusively) circulating in birds [8, 9]. Most CoVs are disease-causing agents, associated with a range of respiratory, gastrointestinal, hepatic and neurological diseases both in humans and animals. They are also potentially transmitted among species, causing epidemics after host transition [10–12]. Due to mutation and recombination events, new strains constantly originate and spread [4, 13].
Different bat species have been identified as natural reservoirs of most mammalian α-CoV and β-CoV, including the Severe Acute Respiratory Syndrome SARS-CoV, and the Middle East Respiratory Syndrome MERS-CoV [14, 15], which recently caused outbreaks in the human population [4]. SARS-related CoV was first identified in Rhinolophus bats from China [16], and subsequently many mammalian CoVs were detected in Chinese bats [17, 18] and globally [19]. Further investigations identified α-CoV and β-CoV in several European countries from bats of various species [20–22].
In Italy, few studies have been conducted on bat CoV ecology: viral detection and prevalence was investigated in greater horseshoe bats [23], alpha- and betacoronaviruses were identified in different species, such as Kuhl’s pipistrelle, lesser horseshoe bats and serotine bats [10, 24], and recently two MERS related beta CoV detected in Italian Kuhl’s pipistrelle and Savi’s bat were fully sequenced [25]. A total of 21 bat species are currently recognized as living on the island of Sardinia (Italy), lying geographically at the interface between Europe and Africa, and recently indicated as a major Mediterranean hotspot for bat biodiversity [26]. Nothing is known about CoV presence and distribution among bat populations on the island, and this is the first study aiming to fill this gap. Herein we report the molecular identification and typing of a SARS-like Coronavirus detected in three species of Sardinian bats belonging to Rhinolophus, Plecotus and Tadarida genera, during a viral survey conducted on non-invasive bat samples collected from different localities on the island. We also inferred the phylogenetic characteristics of the identified β-CoV, and its evolutionary relationships with other bat CoVs identified in Europe and globally.
Materials and methods
Bat sample collection
A total of 46 bats, belonging to 15 different species, were captured using mist nets or hand nets in 8 different localities distributed in northern and central Sardinia: Grotta sa Rocca Ulari (Borutta), Galleria Casteldoria (Santa Maria Coghinas), Diga Santa Chiara (Ula-Tirso), Monte Corallinu (Dorgali), Tilìpera (Bonorva), Mularza Noa (Bolotana), Belvì, and Grotta Verde (Alghero), listed in Table 1. Sampling locations included caves, buildings, tunnels, woods, abandoned bridges, and other type of natural roosts. Captures were performed by expert and trained biologists within a regular census program of Sardinian bat populations, between November 2015 and November 2016. Bats were placed individually into cotton bags before investigation. Species, sex, age category, and forearm length were determined; oral and cutaneous swabs were collected using sterile micro-swabs (Aptaca), and then bats were released at their capture site. Faecal samples were readily collected from cotton bags or during bat handling and suspended in 1 ml RNA later RNA stabilization solution (Qiagen) and then stored at − 20 °C before processing.
Table 1.
List of bat samples analyzed: host species, sampling locations, and type of samples collected
| Host species | Sampling location | Skin swab | Oral swab | Faeces |
|---|---|---|---|---|
| Rhinolophus mehelyi | 1 | 2 | – | 2 |
| Rhinolophus ferrumequinum | 1, 2, 8 | 3 | 3 | 3 |
| Miniopterus schreibersii | 1 | 9 | – | 7 |
| Myotis punicus | 1, 2, 5 | 8 | 8 | 6 |
| Myotis capaccinii | 2, 8 | 3 | 4 | 3 |
| Myotis emarginatus | 2, 6 | 3 | 2 | 2 |
| Plecotus sardus | 3 | – | 1 | – |
| Hypsugo savii | 4, 6 | 1 | 1 | – |
| Pipistrellus pipistrellus | 5 | 2 | – | – |
| Pipistrellus kuhlii | 6 | – | – | 1 |
| Plecotus auritus | 7 | 1 | 1 | 1 |
| Plecotus austriacus | 8 | – | 1 | – |
| Nyctalus leisleri | 6 | 1 | 1 | – |
| Eptesicus serotinus | 8 | – | – | 1 |
| Tadarida teniotis | 8 | 1 | 1 | 1 |
| Tot | 34 | 23 | 27 |
Locations: (1) Grotta sa Rocca Ulari (Borutta), (2) Galleria Casteldoria (S.M. Coghinas), (3) Diga S.Chiara (Ula-Tirso), (4) Monte Corallinu (Dorgali), (5) Tilipera (Bonorva), (6) Mularza Noa (Bolotana), (7) Belvì, (8) Grotta Verde (Alghero)
DNA and RNA extraction, reverse transcription
DNA was extracted from oral and skin swabs tips using the QiAmp DNA Mini Kit (Qiagen), following the manufacturer’s protocol specific for swabs. DNA was stored at − 20 °C for subsequent amplification analysis. After suspending faecal samples in phosphate buffered saline (PBS), RNA was extracted from faeces by using the QiAmp Viral RNA Mini Kit (Qiagen), according to the instructions suggested by the manufacturer. Extracted RNA was stored at − 80 °C. First-strand cDNA was synthesized utilizing the Affinity Script Multi Temperature cDNA Synthesis Kit (Agilent Technologies), following the random primers strategy protocol, and then stored at − 20 °C.
PCR and RT-PCR, screening and sequencing
DNA extracted from bat oral swabs was tested for the presence of Herpesvirus using a universal nested PCR [27]. DNA extracted from skin swabs was screened for Poxvirus and Papillomavirus presence, by PCR based on pan-pox primers [28] and on primers targeting a conserved region of PPV L1 gene [29].
RT-PCR was performed to detect Coronavirus in bat faecal samples, by using two pairs of primers targeting a conserved fragment of RNA-dependent RNA polymerase gene (RdRp): 11FW and 13RV [23], and Corona 1 forward and Corona 2 reverse [10, 17, 30], targeting 170 bp and 440 bp respectively. In a typical PCR, 2 µl of cDNA was amplified in a 50 µl reaction mixture containing 0.2 mM of each dNTPs, 0.2 µM each of Forward and Reverse primers, 1X TaqBuffer and GoTaq (Promega, Italy) DNA polymerase. The cycling conditions were as follows: 5 min at 95 °C followed by 40 cycles of 1 min at 95 °C, 1 min at 50 °C, and 1 min at 72 °C, followed by a final extension time of 5 min at 72 °C. From the first amplified sequence of approximately 170 bp, in order to further amplify the RdRp gene, 4 sets of primers were newly designed, after aligning the sequence detected in this study with the most similar sequences obtained by BLAST analysis, and choosing conserved regions to design consensus walking primers. The first 170 bp amplicon was extended by designing and using downstream primers within the fragment and upstream primers based on alignments of phylogenetically related CoV from GenBank. Table 2 lists the primers designed in this study (BatCV1F/R, BatCV2F/R, BatCV3F/R, and BatCV4F/R). PCR products were purified using DNA Clean & Concentrator purification kit (Zymo Research, CA, USA), and directly sequenced in both forward and reverse directions by BMR Genomics Sequencing Service (Padova, Italy). Forward and reverse sequences were edited and aligned with MUSCLE Multiple Sequence Alignment tool (http://www.ebi.ac.uk/Tools/msa/), alignments were edited with BioEdit software [31]. Sequences were checked against the GenBank database using nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 2.
Primer sequences used and designed in this study for bat Coronavirus detection
| Name | Target sequence | Primer sequence | Source |
|---|---|---|---|
|
11FW 13RV |
RdRp |
5′-TGATGATGCCGTCGTGTGCTACAA-3′ 5′-TGTGAGCAAAATTCGTGAGGTCC-3′ |
Balboni et al. [23] |
|
BatCV1F BatCV1R |
RdRp |
5′-TACAGTGATGTAGAAACTCC-3′ 5′-AAGGTCTGTCTCAGTCCAGCA-3′ |
This study |
|
BatCv2F BatCV2R |
RdRp |
5′-TGCTGGACTGAGACAGACCTT-3′ 5′-GTGTTATCATTAGTTAGCATT-3′ |
This study |
|
BatCV3F BatCV3R |
RdRp |
5′-CTAAGCGCAATGTCATCCCT-3′ 5′-GCATGTTAGGCATGGCTCT-3′ |
This study |
|
BatCV4F BatCV4R |
RdRp |
5′-GCATCAAGAACTTTAAAGCAG-3′ 5′-GAATACATGTCTAACATGTG-3′ |
This study |
Phylogenetic analysis
Phylogenetic trees were estimated using the Maximum Likelihood (ML) and Maximum Parsimony (MP) methods implemented in MEGA 6 [32], after testing the best fitting evolutionary model to the data. Bootstrap support values and reliability of internal branches were calculated from 1000 replicate trees. Models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. Non-uniformity of evolutionary rates among sites was modeled by using a discrete Gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionary invariable (+I). After generating a gap-free nucleotide alignment of 1090 bp containing the novel virus as well as of Coronavirus RdRp reference strains, the most likely nucleotide substitution model selected to analyze these data was TN93+G (Tamura-Nei, Gamma distributed), while analyzing an alignment of a higher number of CoV RdRp sequences of 396 bp, the model T92+G was selected (Tamura 3-parameter, Gamma distributed). The CoV sequence obtained in this study was submitted to the GenBank database under accession number MG978754.
Results
Coronavirus RNA was detected in three out of 27 faecal samples: 11%, close to previously reported frequency [10], less than that observed in other studies [17, 30], unlike results obtained in central-southern Italy [33]. Positive samples were collected in different locations of northern and central Sardinia from three different bat species: the greater horseshoe bat (Rhinolophus ferrumequinum), the brown long-eared bat (Plecotus auritus), and the European free-tailed bat (Tadarida teniotis). Table 3 lists samples positive for CoV detected in this study, including the GenBank accession number assigned to the novel sequence. No Herpesvirus, Poxvirus and Papillomavirus were detected in the oral and/or cutaneous samples collected within this Sardinian bats viral survey.
Table 3.
List of bat positive samples for Coronavirus, and GenBank accession number
| ID | Host species | Sample type | Sex | Location | CoV strain | GenBank accession number |
|---|---|---|---|---|---|---|
| 15F | Greater Horseshoe bat R. ferrumequinum | Faeces | F | 2 | SarBatCoV1 | MG978754 |
| 30F | Brown Long-eared bat P. auritus | Faeces | nd | 7 | SarBatCoV1 | – |
| 31F | European Free-tailed bat T. teniotis | Faeces | F | 8 | SarBatCoV1 | – |
Locations (ref. Table 1): 2. Galleria Casteldoria (S.M. Coghinas), 7. Belvì, 8. Grotta Verde (Alghero)
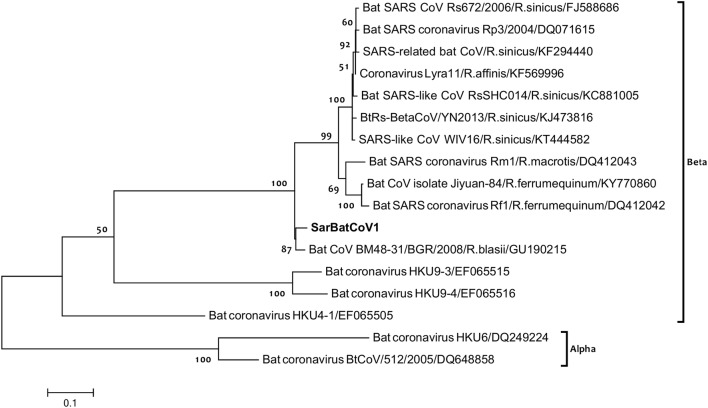
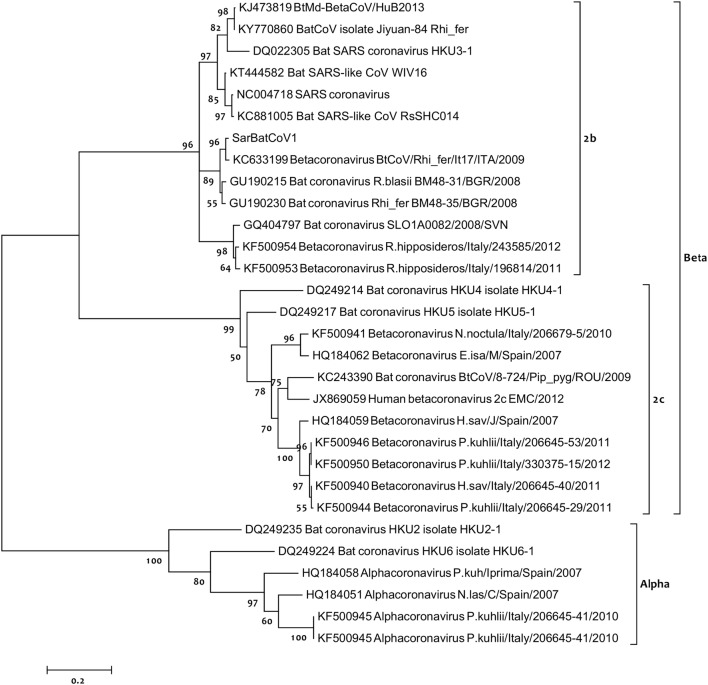
After detecting CoV in faecal samples, a total of 1110 bp of RdRp gene were successfully amplified and directly sequenced. The same betacoronavirus sequence was detected in all three positive bat species. Upon BLASTN analysis, the viral sequence identified was found highly similar to Bat coronavirus BM48-31/BGR/2008 (GU190215), detected in a Blasius’s horseshoe bat (Rhinolophus blasii) in Bulgaria [14], with 100% query coverage and 96% identity. The obtained sequence also showed high nucleotide similarity (89%) with Bat SARS CoV Rs672/2006 (FJ588686) identified in a Chinese rufous horseshoe bat (Rhinolophus sinicus) [34], and with Bat SARS-like coronavirus RsSHCO14 (KC881005) from the same bat species [35]. A first phylogenetic analysis was performed based on 1090 bp amplified fragment of the RdRp gene including the CoV identified in Sardinian bats and other 16 sequences representatives of CoV genetic variability. The phylogram (TN93+G, Fig. 1) clearly showed that Sardinian bat CoV belong to the β-CoV genus, and clustered together with the Bulgarian bat CoV identified in a Rhinolophus blasii (GU190215). To have a deeper look into clade structuring within genera and further characterize the phylogenetic relationships of the detected CoV, especially with other European bat coronaviruses, another phylogenetic analysis was performed based on 396 bp of the RdRp gene, and including a total of 30 sequences. Figure 2 shows the (T92+G) tree obtained, clearly assigning the Sardinian bat CoV to clade 2b within the β-CoV genus. The branching also showed that the detected sequence was closely related to a betacoronavirus previously identified in a greater horseshoe bat (Rhinolophus ferrumequinum) in Italy (BtCoV/Rhi_fer/It17/ITA/2009, GenBank number KC633199), as well as other bat CoVs detected in Rhinolophus spp. from Slovenia (GQ409747) [36], and again from Italy (i.e., KF500954) [10]. All other clade groupings were consistent with previous studies [14].
Fig. 1.
Phylogenetic analysis based on 1090 bp of the RdRp gene. RNA-dependent RNA polymerase-based phylogeny including the novel bat coronavirus detected in this study (SarBatCoV1) and other 16 RdRp sequences from bat coronaviruses. Coronavirus sequence identified in this study is shown in bold and assigned to the β-CoV genus. Bootstrap percentage values from 1000 resamplings are located at nodes of the tree
Fig. 2.
Phylogenetic analysis based on 396 bp of the RdRp gene. Phylogenetic tree of the partial RNA-dependent RNA polymerase gene (396 bp) of coronavirus strains found in bats and humans, showing the assignment of the sequence detected in this study (SarBatCoV1) to clade 2b, within the β-CoV genus. Percentage values from 1000 replicates of bootstrapping are located at nodes of the tree
Discussion
At least 21 bat species are currently recognized as living on the island of Sardinia, Italy [37]. Some species are common and adaptable, inhabiting both wild and urban habitats, while others are rare and restricted to limited areas of the island. Recently an island endemic species was characterized (Plecotus sardus) and an endemic lineage of Plecotus auritus was also identified [38]. The existence of endemic bat taxa on Sardinia highlights the island importance in the conservation of the European bat community. Some Sardinian bat populations appear genetically differentiated and isolated from any continental gene pool and should therefore be considered as an evolutionarily significant unit (ESU) [39].
This is the first report of Coronavirus detection in faeces from bat species in Sardinia. A SARS-like betacoronavirus was detected in three species (Table 3): the greater horseshoe bat (Rhinolophus ferrumequinum), common cave species distributed all over the island; the brown long-eared bat (Plecotus auritus), rare species inhabiting few localities (especially mountain woods) in central Sardinia; and the European free-tailed bat (Tadarida teniotis), common species usually colonizing rocky slopes and Mediterranean bushes as well as buildings and bridges in urban areas.
The detected virus (SarBatCoV1) belonged to clade 2b, including most of the SARS-like CoVs found in bats, as well as human SARS Coronavirus (Figs. 1, 2). SarBatCoV1 appeared related to another betacoronavirus previously identified in the Italian greater horseshoe bat (BtCoV/Rhi_fer/It17/ITA/2009, GenBank number KC633199), and also clustered with a β-Coronavirus isolated from a Blasius’s horseshoe bat in Bulgaria (BM48-31/BGR/2008, GenBank number GU190215). As shown in Fig. 2, the two clades within β-Coronaviruses are strongly supported in the phylogram, and clade 2c clustered many other bat CoVs identified in Europe (Italy and Spain) from different species. These findings confirm a correlation between SARS-like CoVs and bats belonging to Rhinolophus spp., consistently with previous studies [14, 40], although the same RdRp sequence, in this study, was also detected in two other species belonging to the genera Plecotus and Tadarida. As previously discussed, phylogenetic analyses providing insights into viral transmission among bat species have revealed a general clustering by geographic location rather than by bat species [4]. However, our results also show that similar coronaviruses (with nucleotide identity up to 96%) are found in bats distributed in geographically distant regions, such as the island of Sardinia, mainland Italy and Bulgaria.
Since the identification of SARSr-Rh-BatCoV in bats of the Rhinolophus genus [16], an intense effort was dedicated to investigate CoVs in bats, and many SARS-like viruses were detected and characterized in various species worldwide [4, 19]. Recent studies also provided evidence that bats may be the original ancestor hosts for SARS and MERS viruses, which recently caused pandemic and heavy consequences in humans [41–43]. Therefore, it is paramount to investigate and characterize CoVs distribution and diversity in bat species, before their potential emergence in humans.
Full-length genome analysis of the detected CoV should be attempted to thoroughly understand phylogenetic relationships between SarBatCoV1 and betacoronaviruses previously described in European bats. Further studies including larger number of samples as well as continued surveillance on viruses circulating in Sardinian species are also needed to understand the role played by bats in the maintenance and transmission of beta CoVs. Monitoring and research on potential zoonoses are essential, and the prevalence and distribution of CoVs in Sardinian bats should be further investigated in urban and rural environment. However, without specific evidence, bats should not be considered as a major concern for public health, especially given the key ecosystem services they perform [44]. Bats play essential ecological roles in mosquito and pest control, as pollinators, and are keystone species in natural ecosystems. On top of this, bat species are often protected by national and international laws, as their conservation status may be vulnerable or threatened, and Sardinia was recently indicated as a major Mediterranean hotspot for bat biodiversity [26].
Acknowledgements
We thank Luca Montanaro, Gaetano Fichera Enrico Melis, and Manuele Bardi Lecis for their support in the field and technical support.
Author contributions
RL and AA conceived the study; RL conducted field and lab study and analyzed sequence data; MM and EP provided bat samples; RL drafted the manuscript; MP and AA reviewed the ms. All authors revised and approved the manuscript.
Funding
This research was cofunded by the University of Sassari (Italy), PhD course in Life Sciences and Biotechnologies, and Regione Sardegna.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All international, national and institutional guidelines for the care of sampled animals were followed. Samples were collected under ethical and capture permits: “Autorizzazione Ministero Ambiente e Tutela del Territorio e del Mare 0007134 PNM (08/04/2016, DIV II)”, and “Autorizzazione Regione Autonoma della Sardegna, Assessorato Difesa dell’Ambiente, Det. 7325 Rep. N. 171 (13/04/2016)”.
References
- 1.Burgin CJ, Colella JP, Kahn PL, Upham NS. How many species of mammals are there? J Mammal. 2018;99:1–14. doi: 10.1093/jmammal/gyx147. [DOI] [Google Scholar]
- 2.García-Pérez R, Ibáñez C, Godínez JM, Aréchiga N, Garin I, Pérez-Suárez G, de Paz O, Juste J, Echevarría JE, Bravo IG. Novel Papillomaviruses in free-ranging Iberian bats: no virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol Evol. 2014;6:94–104. doi: 10.1093/gbe/evt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith I, Wang L. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 2012;3:1–8. doi: 10.1016/j.coviro.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin XD, Wanga W, Haoc ZY, Wang ZX, Guoa WP, Guana XQ, Wange MR, Wang HW, Zhoua RH, Lia MH, Tang GP, Wug J, Holmesh EC, Zhanga YZ. Extensive diversity of coronaviruses in bats from China. Virol. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse H, Tsang AKL, Tsoi HW, Leung ASP, Ho CC, Lau SKP, Woo PCY, Yuen KY. Identification of a novel bat papillomavirus by metagenomics. PLoS ONE. 2012;7:e43986. doi: 10.1371/journal.pone.0043986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Committee on Taxonomy of Viruses (ICTV) (2017) Taxonomy. https://talk.ictvonline.org/taxonomy
- 10.Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A, Boniotti MB. Detection of coronaviruses in bats of various species in Italy. Viruses. 2013;5:2679–2689. doi: 10.3390/v5112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, Martella V, Cordioli P, Enjuanes L, Buonavoglia C. Recombinant canine Coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J Virol. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decaro N, Mari V, Elia G, Addie DD, Camero M, Lucente MS, Martella V, Buonavoglia C. Recombinant canine Coronaviruses in dogs, Europe. Emerg Infect Dis. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorusso A, Decaro N, Schellen P, Rottier PJM, Buonavoglia C, Haijema BJ, de Groot RJ. Gain, preservation and loss of a group 1a Coronavirus accessory glycoprotein. J Virol. 2008;82:10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Muller MA, Deng H, Herrler G, Drosten C. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of Coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. JVirol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, Drexler JF. Rooting the phylogenetic tree of Middle East Respiratory Syndrome Coronavirus by characterization of a conspecific virus from an African bat. JVirol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 17.Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung YH, Lau SK, Woo PC, Suen KY, Yuen KY, Guan Y, Peiris JS. Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falcón A, Vázquez-Morón S, Casas I, Aznar C, Ruiz G, Pozo F, Perez-Breña P, Juste J, Ibáñez C, Garin I, Aihartza J, Echevarría JE. Detection of alpha and betacoronaviruses in multiple Iberian bat species. Arch Virol. 2011;156:1883–1890. doi: 10.1007/s00705-011-1057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloza-Rausch F, Ipsen A, Seebens A, Göttsche M, Panning M, Drexler JF, Petersen N, Annan A, Grywna K, Müller M, Pfefferle S, Drosten C. Detection and prevalence patterns of Group I Coronaviruses in bats, Northern Germany. Emerg Infect Dis. 2008;14:626–631. doi: 10.3201/eid1404.071439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reusken CBEM, Lina PHC, Pielaat A, de Vries A, Dam-Deisz C, Adema J, Drexler JF, Drosten C, Kooi EA. Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe. Vect Borne Zoonotic Dis. 2010;10:785–791. doi: 10.1089/vbz.2009.0173. [DOI] [PubMed] [Google Scholar]
- 23.Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M (2012) A real-time PCR assay for bat SARS-like Coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci World J 2012:989514 [DOI] [PMC free article] [PubMed]
- 24.De Benedictis P, Marciano S, Scaravelli D, Priori P, Zecchin B, Capua I, Monne I, Cattoli G. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes. 2014;48:366–371. doi: 10.1007/s11262-013-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno A, Lelli D, de Sabato L, Zaccaria G, Boni A, Sozzi E, Prosperi A, Lavazza A, Cella E, Castrucci M, Ciccozzi M, Vaccari G. Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy. Virol J. 2017;14:239. doi: 10.1186/s12985-017-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veith M, Mucedda M, Kiefer A, Pidinchedda E. On the presence of pipistrelle bats (Pipistrellus and Hypsugo; Chiroptera: Vespertilionidae) in Sardinia. Acta Chiropter. 2011;13:89–99. doi: 10.3161/150811011X578642. [DOI] [Google Scholar]
- 27.VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for Poxvirus detection. J Clin Microbiol. 2010;48:268–276. doi: 10.1128/JCM.01697-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Tris J, Williams RAJ, Abel-Fernandez E, Barreiro J, Conesa JJ, Figuerola J, Martinez-Martinez M, Ramirez A, Benitez L. A multiplex PCR for detection of poxvirus and papillomavirus in cutaneous warts from live birds and museum skins. Avian Dis. 2011;55:545–553. doi: 10.1637/9685-021411-Reg.1. [DOI] [PubMed] [Google Scholar]
- 30.De Sales Lima FE, Campos FS, Kuner Filho HC, De Carvalho Ruthner Batista HB, Carnielli PJ, Cibulski SP, Spilki FR, Roehe PM, Franco AC. Detection of Alphacoronavirus in velvety free-tailed bats (Molossus molossus) and Brazilian free-tailed bats (Tadarida brasiliensis) from urban area of Southern Brazil. Virus Genes. 2013;47:164–167. doi: 10.1007/s11262-013-0899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amoroso MG, Russo D, Lanave G, Cistrone L, Pratelli A, Martella V, Galiero G, Decaro N, Fusco G. Detection and phylogenetic characterization of Astroviruses in insectivorous bats from Central-Southern Italy. Zoonoses Public Health. 2018 doi: 10.1111/zph.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan J, Hon CC, Li Y, Wang D, Xu G, Zhang H, Zhou P, Poon LLM, Lam TTY, Leung FCCL, Shi Z. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol. 2010;91:1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- 35.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rihtaric D, Hostnik P, Steyer A, Grom J, Toplak I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch Virol. 2010;155:507–514. doi: 10.1007/s00705-010-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mucedda M, Pidinchedda E (2010) Pipistrelli in Sardegna. Conoscere e tutelare i mammiferi volanti. Nuova Sampa Color, Muros: 1–46
- 38.Mucedda M, Kiefer A, Pidinchedda E, Veith M. A new species of long-eared bat (Chiroptera, Vespertilionidae) from Sardinia, Italy. Acta Chiropterolog. 2002;42:121–135. doi: 10.3161/001.004.0202. [DOI] [Google Scholar]
- 39.Biollaz F, Bruyndonckx N, Beuneux G, Mucedda M, Goudet J, Christe P. Genetic isolation of insular populations of the Maghrebian bat, Myotis punicus, in the Mediterranean Basin. J Biogeogr. 2010 doi: 10.1111/j.1365-2699.2010.02282. [DOI] [Google Scholar]
- 40.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, Ferreira da Silva Filho LVR, Leroy EM, Thiel V, van der Hoek L, Poon LLM, Tschapka M, Drosten C, Drexler JF. J Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S (2017) Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol JVI-01953-16 [DOI] [PMC free article] [PubMed]
- 44.Lòpez-Baucells A, Rocha R, Fernàndez-Llamazares A. When bats go viral: negative framings in virological research imperil bat conservation. Mammal Rev. 2017 doi: 10.1111/mam.12110. [DOI] [Google Scholar]