Abstract
Fecal samples (n = 107) were collected from cattle with ascertained or suspected diarrheal disease on Korean farms during 2008–2010. Of these, 37 samples tested positive for bovine kobuvirus. The 37 positive samples came from 32 cattle that exhibited diarrhea and five cattle that were non-diarrhetic. The majority of the virus-positive feces samples were from calves under 1 month of age (n = 25). Nine of the 37 cattle infected with bovine kobuvirus were confirmed to have a co-infection with other viruses including bovine rotavirus (n = 3), bovine coronavirus (n = 1), bovine viral diarrhea virus (n = 1), and both bovine coronavirus and bovine viral diarrhea virus (n = 4). A neighbor-joining tree grouped 36 of the Korean kobuvirus strains (with the exception of the KB8 strain) into three clusters (G1, G3, and G4), while strains derived from Thailand and Japan (except the U1 strain) were included in the G2 cluster. The results indicated that Korean bovine kobuvirus has diverse lineages regardless of disease status and species.
Electronic supplementary material
The online version of this article (doi:10.1007/s11262-011-0593-9) contains supplementary material, which is available to authorized users.
Keywords: Cattle, Kobuvirus, Phylogeny
Picornaviruses are non-enveloped, single-stranded RNA viruses with a positive sense polarity. They cause a variety of important diseases in humans and animals [1]. The genus Kobuvirus (family Picornaviridae) includes genome ranging in length from 8.2 to 8.3 kb. The polyprotein of kobuvirus is cleaved post-translationally into three structural viral proteins (VP0, VP1, and VP3, with VP0 apparently not being cleaved further) and seven nonstructural proteins (2A–2C and 3A–3D). Kobuviruses encompass two officially recognized species, Aichi virus and bovine kobuvirus, as well as one candidate species, porcine kobuvirus [2–4]. Aichi virus was first isolated in 1991 from a person with acute gastroenteritis in Japan [2]. Bovine kobuvirus (strain U-1) was detected from serum and fecal samples of clinically healthy cattle in Japan [3]. It was subsequently isolated from cattle fecal samples in Thailand [5], Hungary [6], and Belgium [7]. In addition, bovine kobuvirus has been recently detected in domestic sheep in Hungary [8].
This study was performed to further characterize Korean bovine kobuvirus and to analyze the phylogenetic relationship between Korean strains and strains of other origin.
A total of 107 fecal samples were collected from cattle with ascertained or suspected diarrheal disease at 107 farms throughout Korea from January 2008 to July 2010. The cattle included 61 calves under the age of 30 days and 46 cows over the age of 30 days; Out of the total 107 fecal samples, 86 were from animals with diarrhea and 21 from non-diarrheic animals. The cattle were composed of 78 Korean cattle and 29 Holstein cattle. Samples were collected from the provinces of Gyonggi (n = 32), Gangwon (n = 8), Chungbuk (n = 11), Chungnam (n = 42), Gyongbuk (n = 10), and Gyongnam (n = 4). Viral RNA was extracted from feces using TRIzol LSb according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Bovine kobuvirus was detected from fecal samples using reverse-transcript-polymerase chain reaction (RT-PCR) as previously described [3]. Oligonucleotide primers were designed based on the genome sequence of the U-1 strain (Accession No. AB084788) and have the following sequences: U-1f (sense, 5′-CATGCTCCTCGGTGGTCTCA-3′; nt 7,357) and U-1r (antisense, 5′-GTCCGGGTCCATCACAGGGT-3′; nt 7,987). Together, these primers amplify a 631-bp region of the 3D protein. PCR products of size 631 bp were visualized by electrophoresis and were cloned using the pGEM-T Vector System II (Promega, Madison, WI, USA). The cloned genes (three per sample) were sequenced, using T7 and SP6 promoter-specific primers, with an ABI Prism® 3730xi DNA Sequencer (Applied Biosystems, Foster City, CA, USA) at the Macrogen Institute (Macrogen, Seoul, Korea).
To investigate the relationship between kobuvirus and other bovine viruses that cause diarrhea in cattle, a screening test was conducted using primers specific for the detection of bovine rotavirus (BRV) [9], bovine coronavirus (BCV) [10], and bovine viral diarrhea virus (BVDV) [11], as previously described. Reverse transcription for the extracted RNA was performed using a cDNA synthesis kit (TaKaRa) and random hexanucleotide primers. The RT-PCR was run according to the following temperature–time profile: 42°C for 30 min, then 94°C for 5 min, followed by 35 cycles of virus-specific conditions, as follows: BRV: 94°C for 30 s, 54°C for 1 min and 72°C for 1 min; BCV: 94°C for 1 min, 58°C for 1 min and 72°C for 2 min; and BVDV: 94°C for 1 min, 56°C for 1 min and 72°C for 1 min. For all viruses, the 35 denaturation–annealing–extension cycles were followed by a final extension at 72°C for 10 min. The resulting amplicon sizes were 309 bp for BRV, 730 bp for BCV, and 288 bp for BVDV. The sizes were assessed by 1% agarose gel electrophoresis and confirmed through the sequencing and analysis of the nucleotide sequence of each amplicon.
The nucleotide sequences of the Korean bovine kobuviruses were compared to those of kobuvirus reference strains in the GenBank database by BLAST. The nucleotide sequences were aligned using the Clustal W 1.8x program [12] and.aln files were generated. The.aln files were then converted to.meg files using Mega 4 [13] and a neighbor-joining tree was constructed (bootstrap replicates = 1,000) using the Kimura 2 parameter method for pairwise deletion at uniform rates. The nucleotide sequences of Korean bovine kobuvirus strains were deposited in GenBank as accession numbers HQ650164–HQ650200 (Table 1).
Table 1.
Summary of bovine kobuvirus strains
| Strain | Animal | Agea | Year of isolation | Sample | Country | Province | Co-infection with other pathogensb |
Accession no. |
|---|---|---|---|---|---|---|---|---|
| KB1 | Korean native cattle | 20 D | 2008 | Diarrhea | South Korea | Gangwon | BRV | HQ650164 |
| KB2 | Korean native cattle | 1 M | 2008 | Diarrhea | South Korea | Gangwon | – | HQ650165 |
| KB8 | Korean native cattle | 25 D | 2008 | Diarrhea | South Korea | Chungbuk | BRV | HQ650166 |
| KB9 | Korean native cattle | 25 D | 2008 | Diarrhea | South Korea | Chungbuk | – | HQ650167 |
| KB12 | Korean native cattle | 14 D | 2009 | Diarrhea | South Korea | Gyonggi | – | HQ650168 |
| KB14 | Korean native cattle | 2 M | 2008 | Diarrhea | South Korea | Gyonggi | BCV, BVDV | HQ650169 |
| KBH17 | Holstein | 15 D | 2009 | Diarrhea | South Korea | Chungnam | – | HQ650170 |
| KB18 | Korean native cattle | 14 D | 2009 | Diarrhea | South Korea | Chungnam | BRV | HQ650171 |
| KB31 | Korean native cattle | 14 D | 2010 | Diarrhea | South Korea | Chungnam | BCV , BVDV | HQ650172 |
| KB34 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650173 |
| KB38 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650174 |
| KB39 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650175 |
| KB40 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650176 |
| KB41 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650177 |
| KB42 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650178 |
| KB45 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650179 |
| KB55 | Korean native cattle | 3 M | 2008 | Diarrhea | South Korea | Gyonggi | – | HQ650180 |
| KB59 | Korean native cattle | 3 M | 2008 | Diarrhea | South Korea | Gyonggi | – | HQ650181 |
| KB60 | Korean native cattle | 3 M | 2008 | Diarrhea | South Korea | Chungbuk | – | HQ650182 |
| KB61 | Korean native cattle | 1 D | 2008 | Non-diarrhea | South Korea | Gyonggi | – | HQ650183 |
| KBH62 | Holstein | 1 Y | 2009 | Diarrhea | South Korea | Gyonggi | – | HQ650184 |
| KB65 | Korean native cattle | 3 M | 2008 | Diarrhea | South Korea | Chungnam | BCV, BVDV | HQ650185 |
| KBH67 | Holstein | 1 M | 2008 | Diarrhea | South Korea | Chungnam | BCV, BVDV | HQ650186 |
| KBH71 | Holstein | 1 M | 2008 | Diarrhea | South Korea | Chungnam | – | HQ650187 |
| KBH76 | Holstein | 15 D | 2009 | Diarrhea | South Korea | Chungnam | – | HQ650188 |
| KB83 | Korean native cattle | 1 Y | 2009 | Diarrhea | South Korea | Gyonggi | BCV | HQ650189 |
| KB85 | Korean native cattle | 6 D | 2009 | Non-diarrhea | South Korea | Gyongnam | – | HQ650190 |
| KB86 | Korean native cattle | 2 Y | 2009 | Non-diarrhea | South Korea | Gyongbuk | BVDV | HQ650191 |
| KB87 | Korean native cattle | 2 Y | 2009 | Non-diarrhea | South Korea | Gyongbuk | – | HQ650192 |
| KBH92 | Holstein | 4 M | 2009 | Diarrhea | South Korea | Gyonggi | – | HQ650193 |
| KBH94 | Holstein | 4 M | 2009 | Diarrhea | South Korea | Gyonggi | – | HQ650194 |
| KB95 | Korean native cattle | 4 Y | 2009 | Non-diarrhea | South Korea | Gyongbuk | – | HQ650195 |
| KB96 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650196 |
| KB101 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650197 |
| KB102 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650198 |
| KB104 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650199 |
| KB106 | Korean native cattle | 1 M | 2010 | Diarrhea | South Korea | Chungnam | – | HQ650200 |
| CMB02 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659450 | |
| CMB03 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659451 | |
| CMB06 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659452 | |
| CMB07 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659453 | |
| CMB10 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659454 | |
| CMB11 | Cattle | 7–49 D | 2001–2004 | Stool | Thailand | NT | EF659455 | |
| Z20 | Cattle | 1 Y | 2002 | Stool | Hungary | NT | FJ225406 | |
| N2 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097162 | |
| N5 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097163 | |
| K3 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097152 | |
| K4 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097153 | |
| K6 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097154 | |
| K35 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097155 | |
| K36 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097156 | |
| K38 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097157 | |
| K44 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097158 | |
| K49 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097159 | |
| K55 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097160 | |
| K60 | Cattle | 2–4 Y | 2003 | Stool | Japan | NT | AB097161 | |
| U1 | Cattle | 2003 | Japan | NT | NC_004421 |
aD day, M month, Y year
bBRV bovine rotavirus, BCV bovine coronavirus, BVDV bovine viral diarrhea virus, – no infection, NT not tested
In prior studies, bovine kobuvirus was detected in 12 of 72 (16.7%) stool samples in Japan [3], 6 of 72 (8.3%) fecal samples in Thailand [5], and 2 of 32 (6.25%) fecal samples in Hungary [6]. Korean bovine kobuvirus was markedly more prevalent, being detected in 37 of the 107 (34.6%) fecal samples. Furthermore, the yearly frequency of the Korean positive samples was constant: n = 12 in both 2008 and 2009, and n = 13 in 2010 (Table 1). Thirty-two of the 86 diarrhea samples (37.2%) contained kobuvirus, compared with 5 out of 21 non-diarrhea samples (23.8%). However, this result cannot be taken as evidence of a causal relationship between kobuvirus infection and diarrhea, and such a causal relationship has been questioned in previous analyses [3, 5–7].
Infection by kobuvirus occurred in 38.5% (30 of 78) of the Korean native cattle and 24.1% (7 of 29) of the Holstein cattle. This result indicated that kobuvirus infection is not restricted to a single cattle species. Regarding the age of infected cattle, this study clearly showed a predominance of infection in calves under the age of 1 month (n = 25), a result similar to that of a previous study [5]. Kobuvirus prevalence by geographic region was 45.2% (19/42) in Chungnam, 30% (3/10) in Gyongbuk, 28.1% (9/32) in Gyonggi, 27.3% (3/11) in Chungbuk, 25% (2/8) in Gangwon, and 25% (1/4) in Gyongnam. In spite of these seemingly substantial differences in prevalence, the low numbers of samples did not provide sufficient statistical power to allow any conclusions regarding geographic predilection. The geographic differences can therefore be considered tenuous at this point, requiring further study with larger sample sizes. The clinical significance of any such differences remains unclear.
The combined infection involving bovine kobuvirus and other viruses was observed in nine cattles: BRV (n = 3), BCV (n = 1), BVDV (n = 1), and BCV + BVDV (n = 4). However, it is unclear whether the other viruses are directly associated with the kobuvirus infection. Neighbor-joining analysis revealed that partial nucleotide sequences (590 bp in length) of the 3D genes of 56 bovine kobuvirus (37 from Korea, 13 from Japan, and 6 from Thailand), along with that of the Aichi virus (as the outgroup), fell into four main lineages (G1, G2, G3, and G4). With the exception of the U1 and KB8 strains, all of the sequences fell into one of these four lineages (Fig. 1). The four lineages were supported by high bootstrap values (75–99%) at the node of each branch. Interestingly, the 36 Korean kobuvirus strains formed three lineages (G1, G3, and G4), while the 12 Japanese and six Thailand strains all fell within the G2 lineage (Fig. 1). A future analysis using a larger number of strains may be required to confirm that the U1 and KB8 strains represent the first recognized strains of an additional cluster or two additional clusters.
Fig. 1.
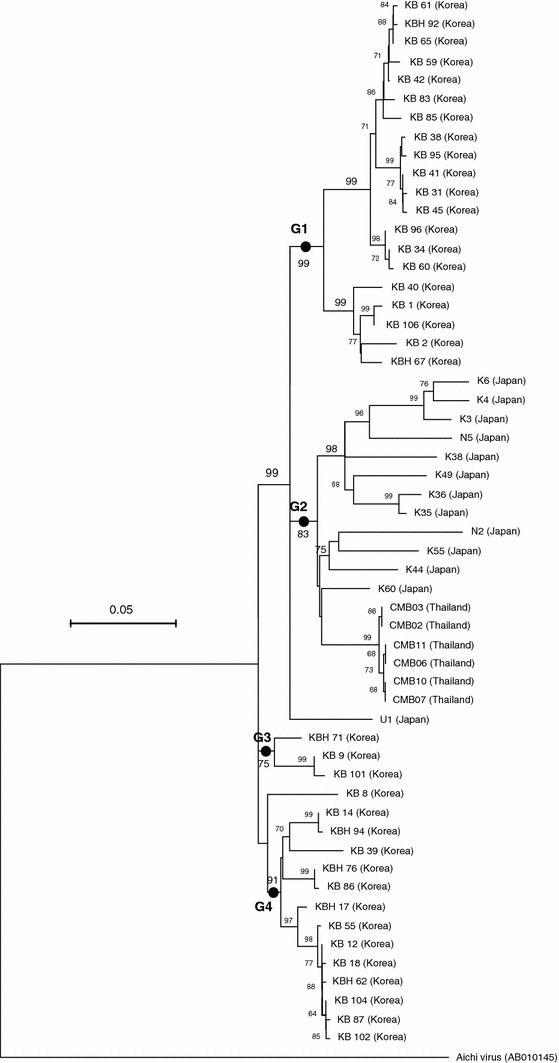
Neighbor-joining tree for nucleotide sequences of bovine kobuvirus strains. Partial nucleotide sequences (590 bp in length) from the 3D genes of 56 global bovine kobuvirus (37 Korean, 13 Japanese, and 6 Thailand), along with the Aichi virus as an outgroup, are shown. Bootstrap percentages are shown above those nodes that are supported in at least 60% of the 1,000 replicates. Scale bar indicates nucleotide substitutions per site
In conclusion, the findings of this study demonstrate the existence of four phylogenetic lineages of bovine kobuvirus. Korean kobuvirus strains are found in three of the four lineages, with Japanese and Thailand strains being clustered together in the other lineage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgment
The authors are grateful to Ms. Bo-Hye Shin and Ms. Hyen-Jung Kim for their technical assistance.
Footnotes
H.-Y. Jeoung and J.-A. Lim contributed equally to this work.
References
- 1.G. Stanway, F. Brown, P. Christian, T. Hovi, T. Hyypiae, A.M.Q. King, N.J. Knowles, S.M. Lemon, P.D. Minor, M.A. Pallansch, A.C. Palmenburg, T. Skern, Picornaviridae. In: Virus Taxonomy, 8th Report of the ICTV, ed. by C.M. Fauquet, M.A. Mayo, U. Desselberger, L.A. Ball, (Elservier Academic Press, Amsterdam, 2005), pp. 757–778
- 2.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. J. Infect. Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. J. Gen. Virol. 2003;84:3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- 4.Reuter G, Boldizsár Á, Kiss I, Pankovics P. Emerg. Infect. Dis. 2008;14:1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khamrin P, Maneekarn N, Peerakome S, Okitsu S, Mizuguchi M, Ushijima H. Emerg. Infect. Dis. 2008;14:985–986. doi: 10.3201/eid1406.070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuter G, Egyed L. Emerg. Infect. Dis. 2009;15:822–823. doi: 10.3201/eid1505.081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauroy A, Scipioni A, Mathijs E, Thys C, Thiry E. Arch. Virol. 2009;154:1841–1845. doi: 10.1007/s00705-009-0518-2. [DOI] [PubMed] [Google Scholar]
- 8.Reuter G, Boros A, Pankovics P, Egyed L. Emerg. Infect. Dis. 2010;16:869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elschner M, Prudlo J, Hotzel H, Otto P, Sachse K. J. Vet. Med. B. 2002;49:77–81. doi: 10.1046/j.1439-0450.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- 10.Cho KO, Hasoksuz M, Nielsen PR, Chang KO, Lathrop S, Saif LJ. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Arch. Virol. 1994;136:309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- 12.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [Google Scholar]
- 13.Tamura K, Dudley J, Nei M, Kumar S. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


